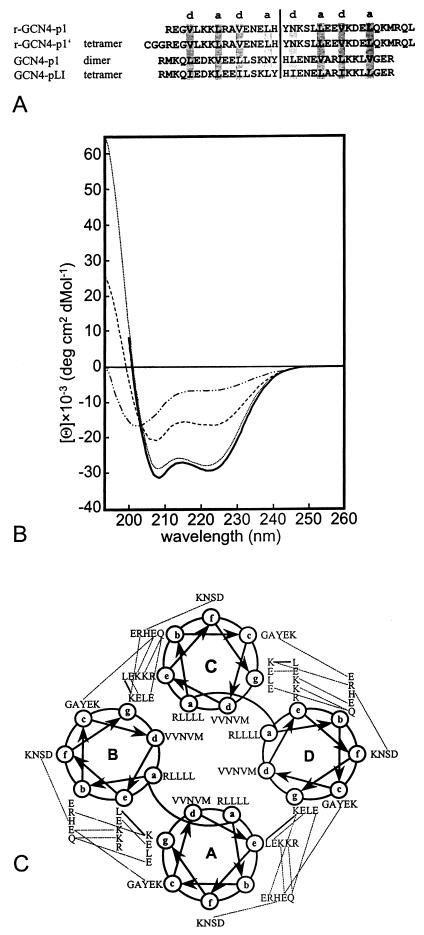
Figure 1.
(A) Sequence alignment of the true 35-residue retro-leucine zipper based on the sequence of GCN4-p1, previously termed r-LZ35 (r-GCN4-p1), r-GCN4-p1′, wild-type C-terminal 33-residue leucine zipper moiety of the yeast transcription activator GCN4 (GCN4-p1), and GCN4-p1 mutant with leucine and isoleucine residues in positions a and d, respectively (GCN4-pLI). Because of the low sequence identity between r-GCN4-p1′ and GCN4-p1 of 19% and the symmetry of the folds, there is no unique superposition. The structures could be shifted by seven residues along the superhelix axis, yielding superpositions with similar rms deviations (rmsds). For this alignment, the conserved central cavity was taken into account. The palindrome axis and the residues that participate in the seven-residue repeats are indicated. Both GCN4-p1 and GCN4-pLI are acetylated at the N terminus. (B) CD spectra of r-GCN4-p1 at various concentrations (1 mM, dotted line; 120 μM, dashed line; 30 μM, dotted/dashed line) and oxidized r-GCN4-p1′ (15 μM, solid line). (C) Helical wheel of the r-GCN4-p1′ tetramer. Helical wheel representation of residues 4 to 36. The view is from the N terminus. Heptad positions are labeled a through g. Polar interactions between side chains are indicated by dashed lines. Main-chain side-chain polar interactions are shown by continuous lines.