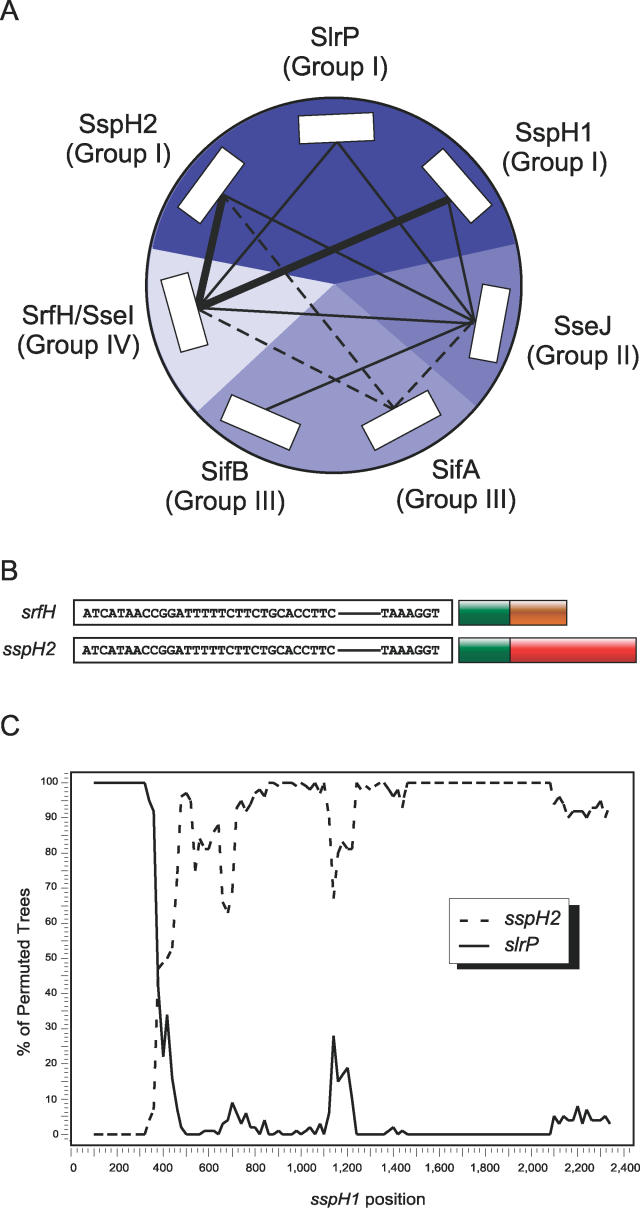
Figure 3. Terminal Reassortment among Effectors in Salmonella enterica.
(A) N-terminal homology among seven effectors of S. enterica representing four distinct homology groups. Each homology group has been encapsulated in a shade of blue and contains proteins that share full-length homology. The interconnecting lines indicate the degree of N-terminal relatedness, with dashed lines indicating <40% similarity, thin, solid lines 40%–60% similarity, and thick, solid lines >60% similarity.
(B) S. enterica effectors sspH2 and srfH share a highly conserved upstream region, ssrAB promoter box, and N-terminal region, while the C terminus of these proteins is non-homologous.
(C) Bootscan of the homologous S. enterica effectors sspH1, sspH2, slrP, and N-terminal homolog sseI/srfH to detect recombination, with the nucleotide sequence of sspH1 as the query sequence. sseI/srfH had <35% permuted trees and was omitted from the figure. Bootscanning was performed with Simplot v 3.5.1 using neighbor joining with the F84 model, gapstrip off, and window and step sizes of 200 and 20, respectively. Other members of this effector family were excluded to facilitate optimal nucleotide alignment.