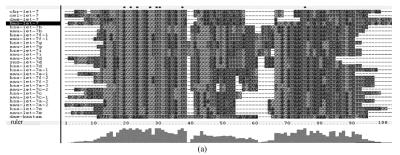
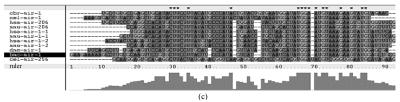
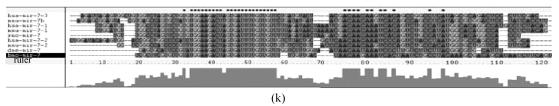
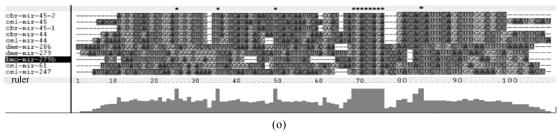
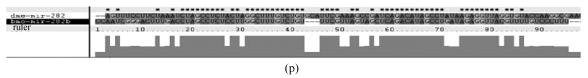
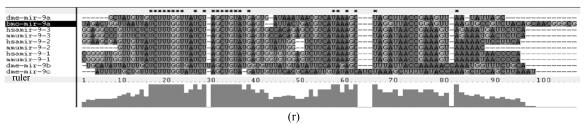
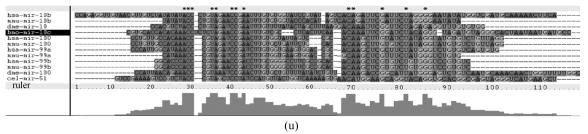
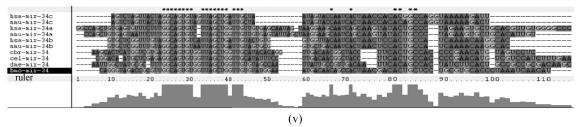
Fig. 1.
Sequence alignments of pre-miRNAs in each miRNA family. (a) let-7 family; (b) mir-124 family; (c) mir-1 family; (d) mir-14 family; (e) mir-275 family; (f) mir-305 family; (g) mir-276 family; (h) mir-263b family; (i) mir-283 family; (j) mir-307 family; (k) mir-7 family; (l) mir-8 family; (m) mir-iab-4 family; (n) mir-31c family; (o) mir-279 family; (p) mir-282 family; (q) mir-263a family; (r) mir-9a family; (s) mir-277 family; (t) mir-71 family; (u) mir-10 family; (v) mir-34 family
Members of each miRNA family that contain novel silkworm miRNAs are subjected to sequence alignment; Precursors of silkworm miRNAs are indicated by black ground. The length of pre-miRNAs consistent with that of foldback predicted sequence. Dark dot in the first line of pictures indicates constant nucleotide