Abstract
Little consideration has been given to the genetic composition of populations associated with marine reserves, as reserve designation is generally to protect specific species, communities or habitats. Nevertheless, it is important to conserve genetic diversity since it provides the raw material for the maintenance of species diversity over longer, evolutionary time-scales and may also confer the basis for adaptation to environmental change. Many current marine reserves are small in size and isolated to some degree (e.g. sea loughs and offshore islands). While such features enable easier management, they may have important implications for the genetic structure of protected populations, the ability of populations to recover from local catastrophes and the potential for marine reserves to act as sources of propagules for surrounding areas. Here, we present a case study demonstrating genetic differentiation, isolation, inbreeding and reduced genetic diversity in populations of the dogwhelk Nucella lapillus in Lough Hyne Marine Nature Reserve (an isolated sea lough in southern Ireland), compared with populations on the local adjacent open coast and populations in England, Wales and France. Our study demonstrates that this sea lough is isolated from open coast populations, and highlights that there may be long-term genetic consequences of selecting reserves on the basis of isolation and ease of protection.
Keywords: Nucella, isolation, marine reserves, genetic diversity, gene flow
1. Introduction
The designation of protected areas is one of the most important tools used in marine conservation, but the main focus of the conservation effort is usually directed towards the protection of a specific species, habitat or biodiversity hotspot, and mostly overlooks genetic diversity (e.g. Kelleher & Kenchington 1991). Marine reserve management and designation have received renewed interest in recent years, with studies focusing on issues such as the persistence of reserve populations, recruitment processes and the potential for self-seeding (Palumbi 2001, 2003; Dethier et al. 2002; Palumbi et al. 2003). However, despite new insights into reserve function and ecology, little attention has focused on whether current marine reserves are suitable for conserving genetic diversity, or the extent that genetic variation of a species is represented in protected populations. Indeed, until recently, the field of conservation genetics has centred primarily on terrestrial ecosystems (Soulé & Terborgh 1999; Pullin 2002), with reserve designation focusing on preserving habitat while maintaining connectivity between populations (Frankham et al. 2003). The direct application of these principles to marine ecosystems is problematic because of the different characteristics these environments display (Carr et al. 2003).
The current marine nature reserves (MNRs) in the UK and Ireland have been designated for a variety of reasons, including long biological histories or recognition as biodiversity hotspots. These sites include sea loughs and offshore islands (Lough Hyne, Strangford Lough, Skomer Island and Lundy Island). As such, marine reserves in the British Isles are small with distinct, easily definable boundaries, and benthic populations found in these sites may be isolated in varying degrees from ‘mainland’ populations. While these characteristics enable effective management, they are likely to make reserve populations more susceptible to environmental change (Hanski 2001), extinction (Frankham 1995, 1998) and inbreeding if the effective population size is small (Wright 1921; Charlesworth & Charlesworth 1987). The level of isolation and the rate of migration between mainland and reserve populations will determine: (i) the amount of buffering the reserve is afforded from local catastrophes by the external input of larvae (Allison et al. 2003); (ii) the usefulness of the reserve as a source of larvae for mainland populations (Chiappone & Sealey 2000); and (iii) the amount of genetic differentiation between reserve and mainland populations.
Marine invertebrates display a spectrum of reproductive strategies that are likely to be a major driving force influencing gene flow and the level of genetic differentiation between populations (Palumbi 1994). The interaction of different life-history traits with environmental and selective forces, as well as species characteristics, must be considered when assessing the potential for genetic isolation in reserve design (Palumbi 1994; Mora & Sale 2002). Many species produce long-lived, free-swimming larvae (planktotrophic) that would normally be expected to disperse widely, linking adult populations across broad spatial scales (Palumbi 2003). Species that produce short-lived larvae (lecithotrophic) may be expected to show lower levels of gene flow between populations (Hoare et al. 1999), while species that lack a larval stage (direct-development) would be expected to show the lowest levels of genetic connectivity. Different levels of genetic disparity might be anticipated between populations of benthic adults with these different reproductive modes. Nevertheless, although long-distance dispersal is predicted for species with long-lived larvae, recent evidence has shown that some long-lived larvae may be retained in the local adult habitat owing to small-scale hydrodynamic processes (Palumbi 2001, 2003; Hellberg et al. 2002). This could mean that larval dispersal might actually be lower for many species than predicted by larval life histories. Therefore, genetic disparity may occur at local (less than 1–10 km) as well as distant (hundreds to thousands of km) scales, even for organisms with the potential for long-distance dispersal (Barber et al. 2000, 2002).
Here, we present a case study designed to assess genetic diversity and connectivity for a population of a common and widespread gastropod in a marine nature reserve. We use the intertidal gastropod Nucella lapillus (L.) as a model species to investigate genetic disparity and gene flow between populations in an isolated sea lough (Lough Hyne MNR) and populations on the nearby open Irish coast, and in England, Wales and France. N. lapillus produces direct-developing larvae (Crothers 1985), and would therefore be expected to show low levels of gene flow and a high degree of genetic differentiation between populations. Lough Hyne MNR is a small sea lough, which is separated from the open Atlantic coast (mainland) by narrow tidal rapids. This isolated sea lough was Europe's first designated marine nature reserve and is a well-known biodiversity hotspot (e.g. Wilson & Picton 1983; Minchin 1987; Bell & Barnes 2000; Maughan & Barnes 2000; Davidson et al. 2004). Our study addresses the following questions: (i) are populations within Lough Hyne genetically distinct from those on the adjacent Irish coastline? (ii) Do levels of genetic differentiation between Lough Hyne populations and those on the nearby Irish coast characterize other populations separated by similar spatial scales (1.5 km)? (iii) Is there evidence of gene flow between Lough Hyne and the Irish coast populations, and how does this compare with populations separated by similar spatial scales (1.5 km)? (iv) Is there evidence for inbreeding within Lough Hyne—as the population size of this species is small and would be expected to have limited dispersal capability? (v) To what extent does Lough Hyne contain populations that are representative of levels of genetic variation within the species? Results of this case study demonstrate marked in situ genetic disparity and isolation associated with an isolated reserve, and thus suggest that genetic isolation should be an important evaluation criterion in present-day and future marine-reserve design, designation and management.
2. Material and methods
(a) Sampling locations
Lough Hyne Marine Nature Reserve (designated 1982; see figure 1a), County Cork, Ireland, is a small (0.8 km2), semi-enclosed sea lough, descending to greater than 50 m depth. This lough is connected to the open Irish coast by narrow (∼25 m wide), shallow (<3 m) tidal rapids (known as the Rapids). A raised sill that runs across the Rapids restricts the tidal flow into Lough Hyne, and during an incoming tide, the water must reach the level of this sill before inflow can begin. This means that inflow of water lasts only 4 h. During inflow, current speeds are very fast (>250 cm s−1) in eastern parts of the lough, decrease westwards across the lough and are negligible in the north basin (Bassindale et al. 1957). Outflow of water lasts approximately 8 h, with very slight water currents (as water is skimmed from the surface in the vicinity of the rapids) in all parts of the lough, with the exception of the Rapids. The movement of most mobile organisms (including larvae) in the lough is likely to be confined to the lough itself because of its size compared with the narrow Rapids, but organisms may move into the lough from the Irish coast during inflow periods.
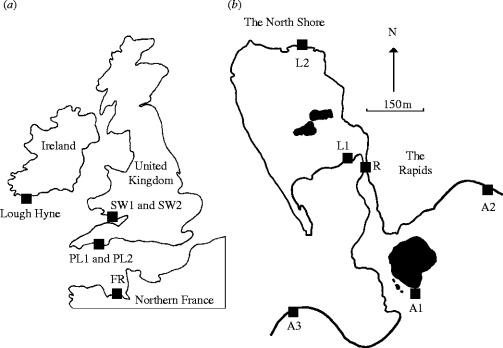
Figure 1.
(a) The sampling location of Nucella lapillus populations around the UK (PL, Plymouth; SW, South Wales; FR, France). (b) The locations where samples were taken within Lough Hyne (L1 and L2), The Rapids (R) and on the surrounding Atlantic coast (A1, A2 and A3).
Thirty N. lapillus were collected from the intertidal zone at two sites (L1 and L2) in Lough Hyne (Renoufs Bay and The North Shore, respectively) and from one site in the Rapids (R; figure 1b). The North Shore and Renoufs Bay intertidal habitats are both very sheltered rocky shores with large boulder areas. The communities at these sites are well developed, but the intertidal zone is limited in vertical extent because the tidal range is only approximately 1.5 m (Bassindale et al. 1957). The population sampled in the Rapids experiences fast water currents (>250 cm s−1) and many of the dogwhelks migrate beneath boulders. Thirty individuals were also collected from three sites (A1, A2 and A3) immediately outside Lough Hyne on the adjacent Irish coast (figure 1b). The first of these sites was at the entrance of the channel connecting the lough to the Atlantic (A1). This site is approximately 1.5 km from the nearest site within Lough Hyne (L1). Sites A2 and A3 were 1.5 km east and west along the shoreline from A1, respectively (figure 1b). These three sites are all characterized by well-developed intertidal communities typical of exposed rocky shores (Ballantine 1961). Populations (n=30) were also sampled at two sites near Plymouth. The first of these sites was at Wembury beach (PL1) and the second site (PL2) was 1.5 km along the coastline towards Fort Bovisand (figure 1a). These sites are also typical, exposed rocky-shore sites (Ballantine 1961). This sampling strategy allowed levels of genetic differentiation between populations to be examined at similar spatial scales to those between Lough Hyne populations and those on the adjacent Irish coast. Two populations (n=30 individuals) were sampled on the south coast of Wales, in the Bristol Channel, at Southerndown (SW1) and Ogmore-by-Sea (SW2) (also 1.5 km apart). Sites SW1 and SW2 were characterized by very sheltered shores, with large sandy areas separated by rocky outcrops covered by Sabellaria reefs. Gastropods were collected from rocky areas between these reefs. Thirty individuals from a single final population (FR) were sampled on the northern French coast (St Malo), which was a moderately exposed rocky shore (figure 1a; Ballantine 1961).
(b) Molecular approaches
Tissue was dissected from the foot of each gastropod and stored in absolute ethanol at 5 °C. DNA was extracted using a CTAB extraction technique (modified from Winnepenninckx et al. 1993) and stored in TE buffer (at −20 °C). Microsatellite markers developed by Kawai et al. (2001) were used to characterize the genetic structure of populations, although polymerase chain reaction (PCR) conditions and annealing temperatures differed. The microsatellites used (with annealing temperature in brackets) were Nlw 3 (55 °C), Nlw 5 (56 °C), Nlw 8 (55 °C), Nwl 11 (54 °C), Nwl 13 (55 °C) and Nwl 14 (54 °C). PCR amplification was carried out in a final volume of 20 μl containing 5 mM Tris–HCl, 10 mM NaCl, 0.01 mM EDTA, 0.1 mM DTT, 5% glycerol, 0.1% Triton X-100, 1.5 mM MgCl2 (PROMEGA storage buffer A), approximately 10 ng of genomic DNA, 0.8 mM dNTPs, 0.5 units of Taq polymerase (PROMEGA) and 0.5 μM of each primer. Thermal cycling (on a Thermohybaid OmN-E) was performed at the following setting: 4 min at 94 °C and 35 thermal cycles, with 30 s at 94 °C, 30 s at optimal annealing temperature (see above) and 30 s at 68 °C and an extra extension step of 8 min at 68 °C. The PCR products were processed using an ABI PRISM 3100 sequencer and alleles for each microsatellite were scored using GENOTYPER (ABI).
(c) Statistical analysis
(i) Genetic diversity
Tests of deviations of genotype frequencies from the Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium were carried out using the exact tests within the computer program GENEPOP (Raymond & Rousset 1995), with significance levels being determined using the Markov chain method (default settings of 100 batches of 1000 iterations). The significance levels of each test were determined by applying the sequential Bonferroni procedure over each loci within each population. Genetic diversity was calculated for each population as the mean number of alleles per locus (A), expected proportion of heterozygotes (HE), observed number of heterozygotes and the inbreeding coefficient (FIS).
(ii) Population genetic structure and differentiation
The genetic structure of populations was assessed using two models: the infinite allele model (Kimura & Crow 1964) and the stepwise mutation model (Ohta & Kimura 1973; Kimura & Ohta 1978). The program Arlequin v. 2.0 (Schneider et al. 2000) was used to calculate values of FST (Weir & Cockerham 1984) and RST (Slatkin 1995) and to conduct analysis of molecular variance (AMOVA). Significance levels were determined for the overall values after 1000 permutations and for population pairwise values after 1000 permutations.
(iii) Isolation and connectivity of populations
Isolation by distance was tested by regressing values of FST and RST with the geographical distance between sites (taken as minimum distance via the marine environment). Mantel tests were used to compare the matrices of pairwise FST and RST and minimum geographical distance using the software programme zt (Bonnet & Van de Peer 2002). This isolation by distance analysis was conducted on all data and on a subset of data that excluded Lough Hyne (L1 and L2) and the Rapids (R) sites. Linear regression analysis was used to examine the relationship between geographical distance between sites and FST/RST. An estimation of the number of migrants per generation (Nm) was calculated as:
where XST is either RST or FST. Assignment tests were performed using the on-line computer program Doh (Brzustowski, J. ‘Doh assignment test calculator’ On-line. Available: http://www2.biology.ualberta.ca/jbrzusto/Doh.php), which is based on the calculations of Paetkau et al. (1995, 1997). This program takes genotypes of individuals from all sampled populations and determines the population from which each individual is most likely to have originated using an assignment index (e.g. Paetkau et al. 1995, 1997). This index can be defined as the highest probability of an individual's genotype in any of the populations. A matrix was constructed (using Doh) of Ax,y, which is a measure of how much more likely are the genotypes of individuals sampled in population x than in population y, using Doh. A similarity dendrogram was produced from these distances using the Drawtree program from the Phylip package (using complete linkage and Canberra distances). Assignment tests were conducted to complement estimation of gene flow via the standard Nm approach described above to avoid the numerous assumptions entailed in derivation of Nm, such as constant population size, an infinite number of populations, constant migration rates and population equilibrium (Whitlock & McCauley 1999).
3. Results
(a) Genetic diversity
The mean number of alleles (table 1) per locus ranged from 3.8 to 8.0, with the lowest number of alleles per locus being found in the two populations within Lough Hyne. The Rapids population had similar numbers of alleles per locus to those populations outside the Lough at local and distant spatial scales. There were no exclusive alleles at any of the sites. Global tests of all data showed a significant departure from HWE. When each population was considered individually, departures from HWE were only found for populations from the two sites within Lough Hyne and for the population in the Rapids (at the 0.05 significance level). Of all the possible comparisons (110 in total), there were only three locus-site pairs that exhibited significant linkage disequilibrium at the 0.05 significance level (Loci Nwl 5 and 13 at sites L1, L2 and R). Independence among loci was therefore assumed. The observed heterozygosity across all loci for each population (table 1) ranged between 0.34 and 0.77. Generally, expected (HE) heterozygosities were only slightly higher than observed (HO) heterozygosities, with the exception of the two populations within Lough Hyne (L1 and L2) and the Rapids. These three populations were characterized by an excess of homozygotes (table 1) with significantly lower HO compared with HE and high inbreeding coefficients (FIS).
Table 1.
Summary of genetic diversity statistics.
| A1 | A2 | A3 | R | L1 | L2 | FR | SW1 | SW2 | PL1 | PL2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 6.8 (±4.1) | 7.6 (±4.5) | 8.2 (±4.7) | 6.4 (±4.3) | 5.0 (±4.3) | 3.8 (±3.0) | 6.9 (±4.1) | 7.6 (±4.3) | 7.8 (±4.6) | 7.8 (±4.0) | 8.0 (±4.6) |
| HO | 0.65 | 0.76 | 0.74 | 0.40 | 0.36 | 0.34 | 0.66 | 0.73 | 0.66 | 0.77 | 0.70 |
| HE | 0.69 | 0.84 | 0.82 | 0.64 | 0.59 | 0.59 | 0.69 | 0.78 | 0.71 | 0.81 | 0.77 |
| FIS | 0.04 | 0.09 | 0.09 | 0.38 | 0.40 | 0.42 | 0.04 | 0.08 | 0.08 | 0.05 | 0.06 |
Mean numbers of alleles per locus (A; ±s.d.), observed heterozygosities (HO), expected heterozygosities (HE) and FIS (inbreeding coefficients) across six microsatellite loci for Nucella lapillus at 11 UK sites (n=30 at each site). Italicized values indicate significant differences (p<0.001) between observed and expected heterozygosities (for a particular site across all loci) and therefore deviation from the Hardy-Weinberg equilibrium. For site descriptions see text.
(b) Genetic differentiation
Significant heterogeneity in genotypic frequency distribution was found in all pairwise comparisons among populations under both the infinite allele (FST) and stepwise mutation (RST) models (p<0.001). Population pairwise comparisons of values of FST were generally smaller than RST. According to Wright's (1978) interpretation of FST values, considerable variation was seen in the degree of genetic differentiation between some pairs of populations. Within-group comparisons (i.e. Irish coast, South Wales, Plymouth, Lough Hyne and French sites) of FST all fell within the 0–0.05 range, indicating little genetic differentiation. Comparisons of FST of populations within Lough Hyne with all other populations showed values of FST between 0.15 and 0.25, indicating a high degree of genetic differentiation. The Rapids population (R) showed only moderate genetic differentiation (FST in the range of 0.05–0.15) to nearby Irish coast populations (A1 and A2) and little differentiation to populations within Lough Hyne. The same patterns pertained to the levels of genetic differentiation based on RST.
A hierarchical analysis of genetic diversity based on variance of allele frequency resulted in significant levels of genetic variance among N. lapillus populations. The AMOVA revealed that nearly 8% of the total microsatellite DNA diversity was explained by the variance among population groups (i.e. Irish coast, South Wales, Plymouth, Lough Hyne and French sites). A smaller proportion of the variance (3.04%) was attributable to within-group population differences, with the largest proportion of the variance being found for the individuals within the total population (89.16%). Each of the three components of the genetic variance was significantly different from zero (p<0.001).
The dendrogram produced by Drawtree (figure 2) indicated considerable genetic differentiation between populations. The Lough Hyne (L1 and L2) and Rapids (R) populations were different from all other populations, and there was relatively high similarity among the Irish coast populations (A1, A2 and A3). These Irish coast populations were still more similar to all other populations (South Wales, Plymouth and France) than to those within Lough Hyne. Little difference was seen between any of the populations separated on small spatial scales (1.5–3 km: SW1 and SW2; PL1 and PL2), with the exception of Lough Hyne compared with the nearby Irish coast populations.
Figure 2.
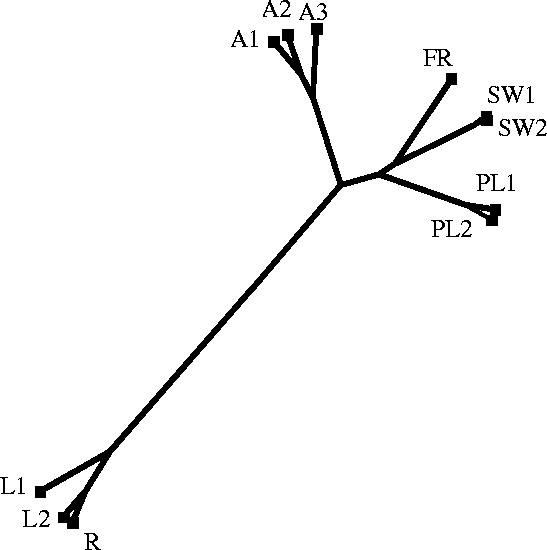
A similarity dendrogram (clustering using Canberra distance and complete linkage) constructed using Drawtree in Phylip based on a distance matrix calculated from assignment indices for populations of Nucella lapillus. A, Atlantic; FR, France; SW, South Wales; PL, Plymouth; L, Lough Hyne; R, Rapids.
(c) Gene flow and individual assignment
Estimated numbers of migrants (Nm) between the pairs of populations varied considerably between sites (table 2), with values based on FST ranging from 0.90 (L1 and A1) to 24.3 (A2 and A3), while values calculated from RST were mostly slightly lower than those calculated from FST values. Generally, populations that were geographically closest had the greatest migration rates (e.g. PL1 and PL2=22.5, SW1 and SW2=20.6, based on FST values). The exceptions were the populations within Lough Hyne and from the Rapids, compared with those immediately outside the Lough on the Irish coast. Although these populations were only separated by 1.5 km (like sites SW1 : SW2 and PL1 : PL2), migration rates were low (0.65–1.5). These estimated migration rates were actually lower than those calculated between the Irish coast populations and populations from all the other sites (Plymouth, France and South Wales). There was evidence for gene flow between the Rapids and Irish coast populations, although greater gene flow was seen between the Rapids and Lough Hyne populations. It was also apparent that greater gene flow occurred between the Rapids and both Lough Hyne populations than seen directly between the two Lough Hyne populations.
Table 2.
Values of RST, FST and Nm.
| A1 | A2 | A3 | R | L1 | L2 | FR | SW1 | SW2 | PL1 | PL2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | — | 0.010 (24.3) | 0.016 (15.4) | 0.187 (1.09) | 0.278 (0.65) | 0.238 (0.80) | 0.079 (2.91) | 0.113 (1.96) | 0.088 (2.59) | 0.133 (1.63) | 0.109 (2.04) |
| A2 | 0.016 (15.4) | — | 0.010 (24.3) | 0.111 (2.0) | 0.184 (1.12) | 0.155 (1.36) | 0.108 (2.06) | 0.070 (3.32) | 0.067 (3.48) | 0.066 (3.54) | 0.067 (3.48) |
| A3 | 0.011 (22.5) | 0.010 (24.3) | — | 0.112 (1.98) | 0.168 (1.24) | 0.143 (1.50) | 0.104 (2.15) | 0.073 (3.17) | 0.080 (2.88) | 0.064 (3.66) | 0.061 (3.85) |
| R | 0.157 (1.34) | 0.099 (2.28) | 0.101 (2.23) | — | 0.036 (6.69) | 0.022 (11.1) | 0.132 (1.64) | 0.118 (1.87) | 0.158 (1.33) | 0.123 (1.78) | 0.121 (1.81) |
| L1 | 0.217 (0.90) | 0.155 (1.09) | 0.144 (1.49) | 0.035 (6.89) | — | 0.066 (3.53) | 0.218 (0.90) | 0.176 (1.17) | 0.244 (0.77) | 0.146 (1.46) | 0.159 (1.32) |
| L2 | 0.192 (1.05) | 0.134 (1.62) | 0.125 (1.75) | 0.021 (11.7) | 0.062 (3.78) | — | 0.194 (1.04) | 0.165 (1.27) | 0.200 (1.00) | 0.135 (1.60) | 0.140 (1.54) |
| FR | 0.074 (3.13) | 0.098 (2.30) | 0.094 (2.41) | 0.117 (1.89) | 0.178 (1.15) | 0.163 (1.28) | — | 0.063 (3.72) | 0.075 (3.03) | 0.102 (2.20) | 0.084 (2.72) |
| SW1 | 0.102 (2.20) | 0.065 (3.60) | 0.068 (3.43) | 0.105 (2.13) | 0.150 (1.42) | 0.141 (1.52) | 0.059 (3.99) | — | 0.012 (20.6) | 0.039 (6.16) | 0.048 (4.96) |
| SW2 | 0.081 (2.84) | 0.063 (3.72) | 0.074 (3.13) | 0.137 (1.57) | 0.196 (1.02) | 0.167 (1.25) | 0.069 (3.37) | 0.012 (20.6) | — | 0.057 (4.14) | 0.061 (3.85) |
| PL1 | 0.117 (1.89) | 0.062 (3.78) | 0.060 (3.92) | 0.108 (2.06) | 0.128 (1.70) | 0.119 (1.85) | 0.093 (2.44) | 0.037 (6.51) | 0.054 (4.38) | — | 0.011 (22.5) |
| PL2 | 0.098 (2.30) | 0.062 (3.78) | 0.060 (3.92) | 0.107 (2.09) | 0.137 (1.57) | 0.123 (1.78) | 0.077 (3.00) | 0.046 (5.18) | 0.058 (4.06) | 0.012 (20.6) | — |
A matrix of pairwise comparisons of population genetic differentiation for Nucella lapillus using RST (infinite allele model) on the lower diagonal and FST (stepwise mutation model) on the upper diagonal. Calculated values of Nm are given in brackets.
Assignment tests provided increased evidence for migration between some populations (table 3). There were considerable numbers of misassigned individuals found between populations that were geographically proximate (e.g. between L1 and L2, SW1 and SW2, PL1 and PL2, Irish coast sites), with the exception of populations inside compared with immediately outside Lough Hyne. There were several misassigned individuals in nearby Irish coast populations, which were originally collected from the Rapids (R), but no individuals found on the Irish coast (A1, A2 and A3) were found to have originated from within Lough Hyne. One individual within the lough was found to have a genotype more similar to those from the Atlantic coast, suggesting unidirectional gene flow. There was also some evidence for longer-distance gene exchange between all pairs of geographically distant populations.
Table 3.
Assignment tests.
| A1 | A2 | A3 | R | L1 | L2 | FR | SW1 | SW2 | PL1 | PL2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 19 | 3 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| A2 | 6 | 21 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A3 | 1 | 4 | 23 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| R | 1 | 0 | 0 | 18 | 6 | 5 | 0 | 0 | 0 | 0 | 0 |
| L1 | 0 | 0 | 0 | 1 | 24 | 5 | 0 | 0 | 0 | 0 | 0 |
| L2 | 0 | 0 | 0 | 7 | 5 | 18 | 0 | 0 | 0 | 0 | |
| FR | 0 | 1 | 0 | 0 | 0 | 0 | 29 | 0 | 0 | 0 | 0 |
| SW1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 20 | 6 | 1 | 1 |
| SW2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 7 | 18 | 2 | 0 |
| PL1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 20 | 7 |
| PL2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 7 | 21 |
The number of misassigned individuals between populations of Nucella lapillus at different northeast Atlantic sites. Values in the table represent the number of assignments from population i (row) to population j (column).
(d) Isolation by distance
Mantel tests indicated no significant correlation between the FST and geographical distance between populations for all the data combined (Mantel test r=0.13, p=0.18, 100 000 permutations). When Mantel tests were used to compare data matrices of geographical distance and RST, the relationship was just significant at the 0.05 level (Mantel test r=0.30, p=0.04, 100 000 permutations). When data from Lough Hyne (L1 and L2) and the Rapids (R) populations were excluded, a significant correlation was found between both FST and RST with the geographical distance between populations (Mantel test r>0.77, p<0.001, 5040 permutations). Linear regression analysis (excluding data from L1, L2 and R) indicated that FST and RST were significantly dependent on geographical distance (F1,27>39.20, p<0.001), with a gradient that was significantly different from zero (t27>6.26, p<0.001).
4. Discussion
(a) Marine reserves, connectivity and isolation
Our results clearly demonstrate that a lack of genetic exchange, inbreeding and reduced genetic diversity characterize a marine-reserve population. This demonstration confirms that degree of isolation may well be an important consideration in reserve management and design, particularly with regard to the long-term sustainability of benthic invertebrate populations in relatively isolated sites, and the ability of such sites to act as sources of propagules for surrounding areas (Palumbi 2003; Shanks et al. 2003). Another potential outcome gained by genetic investigation is the demonstration of genetic distinctiveness of populations. In our case, there was no evidence for unique alleles in the reserve population. Thus, there are a number of ways in which genetic investigations may complement existing biological and ecological considerations to enable effective conservation and management of the marine environment (Palumbi 2003).
It is important to conserve genetic diversity since it provides the raw material for the maintenance of species over longer evolutionary time-scales, and is also of particular relevance at present in terms of providing the basis for responses to rapid environmental change (e.g. climate), since reduced genetic diversity has been correlated with decreased fitness (e.g. Hoelzel et al. 2002). There is no doubt that Lough Hyne populations are smaller and more isolated than populations in most marine reserves, and that these features almost certainly account for the relatively high levels of inbreeding, reduced genetic diversity and isolation for the N. lapillus population reported here. Population size and isolation may be of reduced concern in large-scale reserves, such as occur in the Great Barrier Reef, but isolation versus connectivity remains a relevant issue for networks of marine reserves (Palumbi 2003; Shanks et al. 2003). Thus, although our data may be representative of a relatively extreme situation, they nevertheless provide a case study demonstrating a lack of genetic exchange, inbreeding and reduced genetic diversity in an isolated reserve population. It is therefore of great interest to determine whether such patterns similarly pertain to populations in other existing and proposed marine reserves which are topographically isolated, including islands, sea mounts, bays, lagoons and other sea loughs.
(b) Population genetic structure, gene flow and diversity
Nucella lapillus lacks pelagic larvae and has a maximum lifetime (10 years) migration of 30 m (Crothers 1985; Fretter & Graham 1985). These life-history features should promote mating between close relatives and little gene flow between populations separated by small spatial scales (>1 km). Despite this, we obtained evidence for considerable gene flow (from Nm and assignment tests) between local populations at 1.5 km spatial scales, which is consistent with some other species that lack a long-lived pelagic dispersal phase (Hellberg 1994; Goldson et al. 2001; Duran et al. 2004). The exception was for sites within Lough Hyne compared with the Irish coast. High inbreeding coefficients were also reported for the Lough Hyne and Rapids populations. The reduction in gene flow between populations within the Lough, relative to gene flow between populations at similar scales in other coastal localities, may be explained by the reduced wave action in Lough Hyne. The Rapids population (R) had a more similar genetic structure to the populations within Lough Hyne, although there was some evidence of gene flow with Irish coastal populations. The tidal flow regime during inflow probably explains why the Rapids site exchanged more genetic material with Lough Hyne sites than between Lough Hyne sites (some of the water moves towards L1 and L2, whereas no direct water flow occurs between L1 and L2; Bassindale et al. 1957).
Our study provides strong evidence for genetic isolation between distant populations of N. lapillus when Lough Hyne and Rapids populations were excluded. In contrast, Colson & Hughes (2004) found little evidence for isolation by distance over scales of 10–100 km when studying the recolonization of intertidal sites impacted by Tributyltin (TBT). This chemical was responsible for a widespread increase in imposex (male characteristics are superimposed onto females) and subsequent population extinction of N. lapillus during the 1970s and 1980s (Bryan et al. 1986). The larger spatial scale considered in the present study or the non-equilibrium nature of the newly introduced populations studied by Colson & Hughes (2004) may account for this discrepancy. However, earlier allozyme studies (Day & Bayne 1988; Day 1990; Goudet et al. 1994) and recent microsatellite analysis (Rolàn et al. 2004) have shown significant population differentiation and little gene flow for N. lapillus populations separated by intermediate (kilometres) and small (metres) scales. Such differentiation may reflect adaptation to microscale environmental heterogeneity (Colson & Hughes 2004) as morphological adaptation to different wave exposure regimes has been well documented for this species (Crothers 1985). It is notable that Rolàn et al. (2004) found small (FST=0.025) but significant genetic differentiation between exposed and sheltered populations only 1–2 m apart.
Explanations for the high gene flow observed between some populations in a species lacking pelagic larvae include: rafting as juveniles (e.g. Helmuth et al. 1994; Watts et al. 1998), dislodgement of attached egg capsules through wave action before hatching (J. J. Bell, unpublished observations), crawling larvae (Hellberg 1994) and occasional crawling by adults (Crothers 1985; Fretter & Graham 1985; Marko 1998). Collection of two hatchlings of Nucella emarginata drifting in the intertidal (Martel & Chia 1991) suggests that occasional drifting dispersal may also promote gene flow between populations of N. lapillus.
Historic events may also be important in explaining the present-day diversity patterns observed in the N. lapillus populations. Thus, it is possible that the low genetic diversity in Lough Hyne is at least in part owing to the short geological time since colonization. Although Lough Hyne is thought to have formed by glacial erosion approximately 70 000 years ago, colonization by marine organisms only became possible with the gradual marine transgression of the Lough about 4000 years ago. A combination of historical factors, current low levels of gene flow and local adaptation may therefore explain the genetic disparity of Lough Hyne populations. Whatever the explanation, the genetic disparity of the N. lapillus populations in Lough Hyne contrasts greatly with the rapid attainment of genetic similarity to source populations shown by N. lapillus populations that have re-colonized intertidal areas over the 20-year period following extinction from TBT contamination (Colson & Hughes 2004). Taken together, evidence from these two studies illustrates the population-level consequences of limited (in the case of Lough Hyne) versus high dispersal (TBT areas), and provides a cautionary tale regarding the functional role and location of MNRs. In particular, low levels of genetic diversity that may characterize many resident populations of the Lough Hyne MNR could make them particularly unsuited to responding to environmental change.
Acknowledgments
We thank Matt Henderson, Sylvie Tops and Viv Rimmer for advice and help in microsatellite genotyping, and The School of Animal and Microbial Sciences, University of Reading, for funding. B.O. is grateful for support by a research fellowship from the Leverhulme Trust during the duration of this work. Samples were collected from Lough Hyne with permission from the Irish Heritage Council (Duchas), to whom we are grateful. The authors thank Joanna Freeland for comments on an earlier version of this manuscript. We are most grateful for comments from two anonymous referees that have helped us to significantly improve this manuscript.
Footnotes
Present address: Institute of Biological Sciences, University of Wales, Aberystwyth, Edward Llwyd Building, Aberystwyth, Ceredigion SY23 3DA, UK.
References
- Allison G.W, Gaines S.D, Lubchenco J, Possingham H.P. Ensuring persistence of marine reserves: catastrophes require adopting and insurance factor. Ecol. Appl. 2003;13:8–24. [Google Scholar]
- Ballantine W.J. A biologically-defined exposure scale for the comparative description of rocky shores. Field Stud. Counc. Publ. 1961;1:1–19. [Google Scholar]
- Barber P.H, Palumbi S.R, Erdmann M.V, Moosa M.K. A marine Wallace's line? Nature. 2000;406:692–693. doi: 10.1038/35021135. [DOI] [PubMed] [Google Scholar]
- Barber P.H, Palumbi S.R, Erdmann M.V, Moosa M.K. Sharp genetic breaks among populations of Haptosquilla pulchella (Stomapoda) indicate limits to larval transport: patterns, causes, and consequences. Mol. Ecol. 2002;11:659–674. doi: 10.1046/j.1365-294x.2002.01468.x. [DOI] [PubMed] [Google Scholar]
- Bassindale R, Davenport E, Ebling F.J, Kitching J.A, Sleigh M.A, Sloane J.F. The ecology of Lough Hyne rapids with special reference to water currents. VI. Effects of the rapids on the hydrography of the south basin. Ecology. 1957;45:879–900. [Google Scholar]
- Bell J.J, Barnes D.K. A sponge diversity centre within a marine island. Hydrobiologia. 2000;440:55–64. [Google Scholar]
- Bonnet E, Van de Peer Y. zt: a software tool for simple and partial Mantel tests. J. Stat. Soft. 2002;17:1–12. [Google Scholar]
- Bryan G.W, Gibbs P.E, Hummerstone L.G, Burt G.R. The decline of the gastropod Nucella lapillus around South-West England: evidence for the effect of tributlytin from antifouling paints. J. Mar. Biol. Assoc. UK. 1986;66:611–640. [Google Scholar]
- Carr M.N, Neigel J.E, Estes J.A, Andelman S.A, Warner R.R, Largier J.L. Comparing marine and terrestrial ecosystems: implications for the design of coastal marine reserves. Ecol. Appl. 2003;13:108–116. [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 1987;18:237–268. [Google Scholar]
- Chiappone M, Sealey K.M.S. Marine reserve design criteria and measures of success: lessons learned from the Exuma Cays Land and Sea Park, Bahamas. Bull. Mar. Sci. 2000;66:691–705. [Google Scholar]
- Colson I, Hughes R.N. Rapid recovery of genetic diversity of dogwhelk (Nucella lapillus L.) populations after local extinction and recolonization contradicts predictions from life-history characteristics. Mol. Ecol. 2004;13:2223–2233. doi: 10.1111/j.1365-294X.2004.02245.x. [DOI] [PubMed] [Google Scholar]
- Crothers J.H. Dog-Whelks: an introduction to the biology of Nucella lapillus (L.) Field Stud. 1985;6:291–360. [Google Scholar]
- Davidson I.C, Crook A.C, Barnes D.K.A. Quantifying spatial patterns of intertidal biodiversity: is movement important? Mar. Ecol. 2004;25:15–34. [Google Scholar]
- Day A.J. Microgeographic variation in allozyme frequencies in relation to the degree of exposure to wave action in the dogwhelk Nucella lapillus (L.) (Prosobranchia: Muricacea) Biol. J. Linn. Soc. 1990;40:245–261. [Google Scholar]
- Day A.J, Bayne B.L. Allozyme variations in populations of the dog-whelk Nucella lapillus (Prosbranchia: Muricacea) from the south west peninsula of England. Mar. Biol. 1988;99:93–100. [Google Scholar]
- Dethier M.N, McDonald K, Strathmann R.R. Colonisation and connectivity of habitat patches for coastal marine species distant from source populations. Cons. Biol. 2002;17:1024–1035. [Google Scholar]
- Duran S, Pascual M, Estoup A, Turon X. Strong population structure in the marine sponge Crambe crambe (Peocilosclerida) as revealed by microsatellite marker. Mol. Ecol. 2004;13:511–522. doi: 10.1046/j.1365-294x.2004.2080.x. [DOI] [PubMed] [Google Scholar]
- Frankham R. Inbreeding and extinction: a threshold effect. Conserv. Biol. 1995;9:792–799. [Google Scholar]
- Frankham R. Inbreeding and extinction: island populations. Conserv. Biol. 1998;12:665–675. [Google Scholar]
- Frankham R, Ballou J.D, Briscoe D.A. Cambridge University Press; Cambridge, UK: 2003. Introduction to conservation genetics. [Google Scholar]
- Fretter V, Graham A. The prosobranch molluscs of Britain and Denmark. J. Moll. Stud. 1985;(Suppl. 15) [Google Scholar]
- Goldson A.J, Hughes R.N, Gliiddon C.J. Population genetic consequences of larval dispersal mode and hydrography: a case study with bryozoans. Mar. Biol. 2001;138:1037–1042. [Google Scholar]
- Goudet J, Meeüs T, Day A.J, Gliddon C.J. The different levels of population structuring of the dogwhelk, Nucella lapillus, along the south Devon coast. In: Beaumont A, editor. Genetics and evolution of aquatic organisms. Chapman & Hall; London: 1994. pp. 81–95. [Google Scholar]
- Hanski I. Oxford University Press; Oxford: 2001. Metapopulation ecolgy. [Google Scholar]
- Hellberg M.E. Relationships between inferred levels of gene flow and geographic distance in a philopatric coral, Balanophyllia elegans. Evolution. 1994;48:1829–1854. doi: 10.1111/j.1558-5646.1994.tb02218.x. [DOI] [PubMed] [Google Scholar]
- Hellberg M.E, Burton R.S, Neigel J.E, Palumbi S.R. Genetic assessment of connectivity among marine populations. Bull. Mar. Sci. 2002;70:273–290. [Google Scholar]
- Helmuth B, Veit R.R, Holberton R. Long-distance dispersal of a subantarctic brooding bivalve (Gaimardia trapesina) by kelp rafting. Mar. Biol. 1994;120:421–426. [Google Scholar]
- Hoare K, Hughes R.N, Goldson A.J. Molecular genetic evidence for the prevalence of outcrossing in the hermaphroditic brooding bryozoan Celleporella hyalina (L.) Mar. Ecol. Prog. Ser. 1999;188:73–79. [Google Scholar]
- Hoelzel A.R, Fleischer R.C, Campagna C, Le Boeuf B.J, Alvord G. Impact of a population bottleneck on symmetry and genetic diversity in the northern elephant seal. J. Evol. Biol. 2002;15:567–575. [Google Scholar]
- Kawai K, Hughes R.N, Takenaka O. Isolation and characterisation of microsatellite loci in the marine gastropod Nucella lapillus. Mol. Ecol. Notes. 2001;1:270–272. [Google Scholar]
- Kelleher G, Kenchington R. IUCN; Gland, Switzerland: 1991. Guidelines for establishing marine protected areas. A marine conservation and development report. [Google Scholar]
- Kimura M, Crow J. The number of alleles that can be maintained in a finite allele population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ohta T. Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc. Natl Acad. Sci. USA. 1978;75:2868–2872. doi: 10.1073/pnas.75.6.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko P.B. Historical allopatry and the biogeography of speciation in the prosobranch snail genus Nucella. Evolution. 1998;52:757–774. doi: 10.1111/j.1558-5646.1998.tb03700.x. [DOI] [PubMed] [Google Scholar]
- Martel A, Chia F.S. Drifting and dispersal of small bivalves and gastropods with direct development. J. Exp. Mar. Biol. Ecol. 1991;150:131–147. [Google Scholar]
- Maughan B, Barnes D.K.A. Epilithic boulder communities of Lough Hyne, Ireland: the influences of water movement and sediment. J. Mar. Biol. Assoc. UK. 2000;80:767–776. [Google Scholar]
- Minchin D. Fishes of the Lough Hyne marine reserve. J. Fish Biol. 1987;31:343–352. [Google Scholar]
- Mora C, Sale P.F. Are populations of coral reef fishes open or closed? Trends Ecol. Evol. 2002;17:422–428. [Google Scholar]
- Otha T, Kimura M. The model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a genetic population. Genet. Res. 1973;22:201–204. doi: 10.1017/s0016672300012994. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Calvert W, Sterling I, Strobeck C. Microsatellite analysis of population structure in Canadian Polar Bears. Mol. Ecol. 1995;4:347–354. doi: 10.1111/j.1365-294x.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Waits L.P, Clarkson P.L, Craighead L, Strobeck C. An empirical evaluation of genetic distance statistics using microsatellite data from bear (Ursidae) populations. Genetics. 1997;147:1943–1957. doi: 10.1093/genetics/147.4.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi S.R. Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Syst. 1994;25:547–572. [Google Scholar]
- Palumbi S.R. The ecology of marine protected areas. In: Bertness M, Gaines S.D, Hay M.E, editors. Marine ecology: the new synthesis. Sinauer; Sunderland, MA: 2001. pp. 509–530. [Google Scholar]
- Palumbi S.R. Population genetics, demographic connectivity, and the design of marine reserves. Ecol. Appl. 2003;13:146–158. [Google Scholar]
- Palumbi S.R, Gaines S.D, Leslie H, Warner R.R. New wave: high-tech tools to help marine reserve research. Front. Ecol. Environ. 2003;1:73–79. [Google Scholar]
- Pullin A.S. Cambridge University Press; Cambridge, UK: 2002. Conservation biology. [Google Scholar]
- Raymond M, Rousett F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Rolàn E, Guerra-Varela J, Colson I, Hughes R.N, Rolàn-Alvarez E. Morphological and genetic analysis of two sympatric morphs of the dogwhelk Nucella lapillus (Gastropoda: Muricidae) from Galicia (Northwestern Spain) J. Moll. Stud. 2004;70:179–185. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Genetics and Biometry Laboratory; University of Geneva, Switzerland: 2000. Arlequin ver. 2.000: a software for population genetic analysis. [Google Scholar]
- Shanks A.L, Grantham B.A, Carr M.H. Propagule dispersal distance and the size and spacing of marine reserves. Ecol. Appl. 2003;13:159–169. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulé M.E, Terborgh J. Island; Washington, DC: 1999. Continental conservation. [Google Scholar]
- Watts P.C, Thorpe J.P, Taylor P.D. Natural and anthropogenic dispersal mechanisms in the marine environment: a study using cheilostome bryozoa. Phil. Trans. R. Soc. B. 1998;353:453–464. [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Whitlock M.C, McCauley D.E. Indirect measures of gene flow and migration: FST≠1/(4Nm+1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- Wilson K, Picton B.E. A list of the Opisthobranchia: Mollusca of Lough Hyne Nature Reserve, Co. Cork, with notes on distribution and nomenclature. Ir. Nat. J. 1983;21:69–72. [Google Scholar]
- Winnepenninckx B, Backeljau T, De Wachter R. Extraction of high molecular weight DNA from molecular weight DNA in molluscs. Trends Genet. 1993;9:407. doi: 10.1016/0168-9525(93)90102-n. [DOI] [PubMed] [Google Scholar]
- Wright S. Systems of mating II. The effects of inbreeding on the genetic composition of a population. Genetics. 1921;6:124–143. doi: 10.1093/genetics/6.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Variability within and among populations. vol. 4. Chicago University Press; Chicago, IL: 1978. Evolution and genetics of populations. [Google Scholar]