Abstract
Many Saccharomyces cerevisiae duplicate genes that were derived from an ancient whole-genome duplication (WGD) unexpectedly show a small synonymous divergence (KS), a higher sequence similarity to each other than to orthologues in Saccharomyces bayanus, or slow evolution compared with the orthologue in Kluyveromyces waltii, a non-WGD species. This decelerated evolution was attributed to gene conversion between duplicates. Using ≈300 WGD gene pairs in four species and their orthologues in non-WGD species, we show that codon-usage bias and protein-sequence conservation are two important causes for decelerated evolution of duplicate genes, whereas gene conversion is effective only in the presence of strong codon-usage bias or protein-sequence conservation. Furthermore, we find that change in mutation pattern or in tDNA copy number changed codon-usage bias and increased the KS distance between K. waltii and S. cerevisiae. Intriguingly, some proteins showed fast evolution before the radiation of WGD species but little or no sequence divergence between orthologues and paralogues thereafter, indicating that functional conservation after the radiation may also be responsible for decelerated evolution in duplicates.
Keywords: selective constraints, whole-genome duplication, concerted evolution, decelerated evolution
Gene conversion has been extensively studied in yeast (1, 2). Recently, Kellis et al. (3) identified 60 gene pairs in Saccharomyces cerevisiae that were derived from an ancient whole-genome duplication (WGD) but showed a small sequence divergence. Kellis et al. (3) suggested that these genes have undergone gene conversion for three reasons. First, in 90% of the cases, both paralogues show decelerated evolution (at least 50% slower than the orthologue in Kluyveromyces waltii). Second, nucleotides at fourfold-degenerate codon positions for these genes are highly conserved. Third, in approximately half of the cases, the two paralogues in S. cerevisiae are closer in sequence to each other than either is to its syntenic orthologue in Saccharomyces bayanus. Similarly, Gao and Innan (4) attributed the small synonymous divergence (KS) between ancient duplicated genes in yeast to gene conversion. However, we found that most WGD gene pairs with decelerated evolution (3) have an extremely strong codon-usage bias (Fig. 4, which is published as supporting information on the PNAS web site). Codon-usage bias is known to increase with gene-expression level (5, 6) and can slow down synonymous divergence between duplicate genes (7). Therefore, we investigated whether gene conversion or codon-usage bias was more important for the decelerated evolution.
Results and Discussion
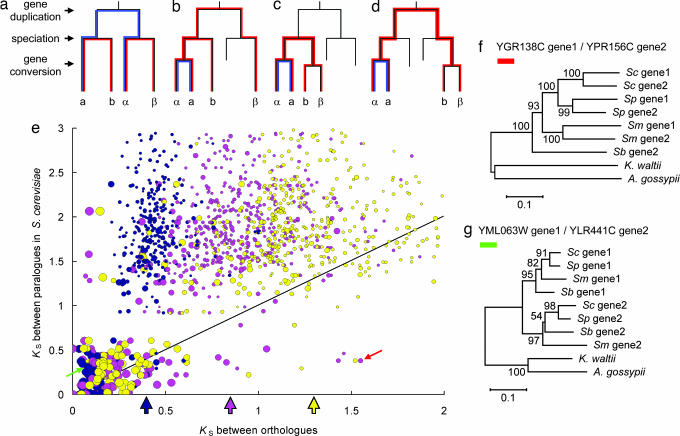
We first use the hypothetical trees in Fig. 1a–d to explain that a gene-conversion event can distort the branch lengths and the topology of the phylogeny of duplicate genes and their orthologues among species. For example, the distance between paralogues α and a is expected to be longer than that between orthologues α and β (Fig. 1a), but the opposite is true in Fig. 1b because of a gene-conversion event. To see how often such a situation has occurred in yeast duplicate genes, we studied ≈300 WGD gene pairs in S. cerevisiae and their syntenic orthologues from three related species, S. bayanus, Saccharomyces mikatae and Saccharomyces paradoxus (8). Because the WGD occurred before the radiation of these species, in the absence of gene conversion, the synonymous distance (KS) is expected to be larger between S. cerevisiae paralogues than between orthologues in different species. We find that this expectation indeed holds in most cases, with 93.4% of duplicate pairs in S. cerevisiae having a paralogous KS greater than or equal to the KS between orthologues (Fig. 1e). This result indicates that only in a small proportion of these WGD duplicate genes has the tree topology been distorted by gene conversion, because only when a point is below the line in Fig. 1e would a distortion in topology have occurred. Interestingly, most S. cerevisiae paralogous pairs with a small KS also show a small KS between orthologues, and many have a high codon-adaptation index (CAI) value (a large circle in Fig. 1e), a measure of codon-usage bias (9). This analysis suggests that decelerated evolution of S. cerevisiae paralogues is, at least in part, due to biased codon usage, which serves as an evolutionary constraint (7, 10).
Fig. 1.
Effects of gene conversion on tree topology and observed patterns of synonymous distances between orthologous or between paralogous genes. (a) Genes a and α (b and β) are paralogues derived from a gene duplication, and a and b (α and β) are orthologues derived from a speciation event. Blue and red lines indicate the distances between paralogues (a and α) and orthologues, respectively. (b) α was converted by a. (c) α was converted by a, and β was converted by b. (d) α was converted by a, and b was converted by β. Note that gene conversion can reduce the distance between paralogues but tends to increase the distance between syntenic orthologues. (e) KS between paralogues in S. cerevisiae (distance between a and α) vs. KS between orthologues (distances between a and b or α and β). Dark blue, S. cerevisiae vs. S. paradoxus; pink, S. cerevisiae vs. S. mikatae; yellow, S. cerevisiae vs. S. bayanus; open arrows indicate the average distances for these species pairs under weak codon-usage bias (KS = 0.4, 0.8, and 1.3). Circle sizes indicate the CAI values of the genes in S. cerevisiae. The slope line indicates that the distance between paralogues is equal to that between orthologues. The red and green solid arrows indicate gene pairs YGR138C/YPR156C between S. cerevisiae and S. mikatae and YML063W/YLR441C between S. cerevisiae and S. paradoxus, respectively. Genes with incomplete sequences, paralogous pairs with KS > 3, and orthologous pairs with KS > 2 are not included in this figure. (f) Neighbor-joining tree (KS distances) of the WGD gene pair YGR138C/YPR156C in S. cerevisiae (Sc), their orthologues in S. paradoxus (Sp), S. mikatae (Sm), and S. bayanus (Sb), and the outgroups in K. waltii (3) and A. gossypii (11) (the red arrow in 1e, CAI = 0.310/0.261). The orthologue of YGR138C in S. bayanus was not completely sequenced and not included in this figure. (g) YML063W/YLR441C (the green arrow, CAI = 0.769/0.696). The numbers at branch nodes are bootstrap values.
We now use two examples to illustrate different effects of gene conversion and codon-usage bias on the evolution of duplicate genes. The first one is the gene pair YGR138C/YPR156C, indicated by the red arrow in Fig. 1e. The small circle indicates that these two genes have a weak codon-usage bias (CAI 0.310/0.261), which is also reflected in the large KS distance between orthologues. However, contrary to expectation, the KS distance between the two S. cerevisiae paralogues is smaller than those between orthologues (Fig. 1e), suggesting that gene conversion has occurred between the two S. cerevisiae paralogues. Indeed, the phylogenetic tree in Fig. 1f shows that the paralogues in each of the first three species are clustered, indicating gene conversions in these species after speciation. The second example is the gene pair YML063W/YLR441C, indicated by the green arrow in Fig. 1e. The large circle indicates a strong codon-usage bias (CAI 0.769/0.696), which is reflected by small KS values between orthologues and between paralogues. The tree topology is as expected (Fig. 1g), so it provides no evidence of gene conversion. Despite this, the tree branches in Fig. 1g are, in general, much shorter than those in Fig. 1f. Clearly, codon-usage bias can slow down sequence evolution in the entire tree, whereas gene conversion can shorten only sequence divergences between paralogues but not those between syntenic orthologues.
To pursue the analysis further, we reconsidered the 66 duplicate gene pairs identified by Gao and Innan (4) to have a small KS between S. cerevisiae paralogues. We found that 57 of them were duplicated before the divergence between S. cerevisiae and S. bayanus, and only one of these 57 pairs (YGL147C/YNL067W) is not from WGD (3, 11). In the 57 phylogenies for these 57 pairs, only 8 pairs showed a completely distorted tree topology (suggesting conversion in all lineages) like Fig. 1f, 23 pairs showed a partially distorted topology, and approximately half of them (26 pairs) showed no topology distortion (Table 3, which is published as supporting information on the PNAS web site). We note that, with the exception of two (YDL131W/YDL182W and YDR312W/YHR066W), all 57 pairs have a strong codon-usage bias (CAI > 0.5). Therefore, in many of these gene pairs, the small KS values between S. cerevisiae paralogues (and between orthologues) might be largely due to strong codon-usage bias constraint.
The above phylogenetic analysis, however, is not powerful enough for detecting all gene-conversion events, because conversion events involving only a small DNA region are unlikely to change the tree topology. For this purpose, we have developed a statistical method to detect gene-conversion events and have applied it to ≈300 WGD duplicate gene pairs in S. cerevisiae, S. paradoxus, S. mikatae, and S. bayanus. Our main purpose is to see whether gene conversion occurred primarily in high-CAI genes. Indeed, Table 1 shows that approximately half of the genes with CAI ≥ 0.7 have undergone gene-conversion events, whereas only 2% of the genes with CAI < 0.5 have conversions (P < 10−8 for all species). Apparently, codon-usage bias increases the rate of gene conversion by reducing the rate of sequence divergence. In the absence of strong codon-usage bias, synonymous divergence between duplicate genes increases with time, and the chance of gene conversion is concomitantly reduced.
Table 1.
Number of gene pairs (with detected gene conversion events/total)
| Species | CAI ≥ 0.7 | 0.7 > CAI ≥ 0.5 | CAI < 0.5 | P |
|---|---|---|---|---|
| S. cerevisiae | 12/21 | 4/31 | 4/238 | <10−8 |
| S. paradoxus | 9/18 | 3/28 | 3/215 | <10−8 |
| S. mikatae | 8/20 | 4/29 | 3/161 | <10−8 |
| S. bayanus | 15/21 | 11/31 | 6/246 | <10−8 |
Only detected conversion events longer than 20 bp were reported.
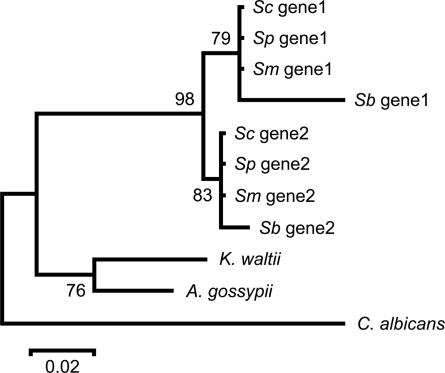
Another intriguing observation was that, for most duplicate gene pairs that show a small protein distance, the divergence between the K. waltii–Ashbya gossypii and Saccharomyces sensu stricto species lineages is much longer (e.g., Fig. 2). This observation has been taken as evidence of gene conversion in the Saccharomyces species under study (3). However, we notice that, in these genes, the protein distances are short not only between paralogues in the same species but also between orthologues in different WGD species, indicating that protein-sequence conservation, rather than gene conversion, was the major cause of decelerated evolution. In the period immediately after the WGD event, the duplicate proteins had apparently evolved rather rapidly (Fig. 2), likely because of relaxed functional constraints after WGD or the emergence of anaerobic growth, which has been found to be connected with cis-regulatory element evolution (12). During this period, gene conversion might have played a key role in maintaining the sequence similarity between the two paralogues. However, the rate of evolution had evidently become very slow before the radiation of the four Saccharomyces species (Fig. 2), largely explaining why the sequence divergence is small between not only paralogues but also orthologues.
Fig. 2.
Neighbor-joining tree of the whole-genome duplicated ORFs of S. cerevisiae (Sc) and their orthologues in S. paradoxus (Sp), S. mikatae (Sm), and S. bayanus (Sb) and outgroups K. waltii, A. gossypii, and Candida albicans for YER131W (gene 1)/YGL189C (gene 2) (cytoplasmic small ribosomal subunits, CAI = 0.711/0.781). The tree was constructed by using protein Poisson distances. The numbers at branch nodes are bootstrap values.
As for synonymous substitutions, previous studies showed that overlooking nucleotide-composition differences (13) or codon-usage patterns (14) among sequences can mislead phylogenetic reconstruction. An examination of the codon-usage patterns reveals that genes in K. waltii and A. gossypii have a stronger preference for G and C at third-codon positions than genes in the four Saccharomyces species (Table 2), perhaps one reason for the large KS values in highly expressed genes between the K. waltii–A. gossypii lineage and the Saccharomyces lineage.
Table 2.
Relative frequencies of codon usage and numbers of tDNA genes for different anticodons in yeast species
| A. gossypii | K. waltii | S. cerevisiae | S. paradoxus | S. mikatae | S. bayanus | |
|---|---|---|---|---|---|---|
| Amino acid–codon | Relative frequencies of codon usage (highly/less expressed genes) | |||||
| Asp–GAU | 0.14/0.44 | 0.16/0.55 | 0.43/0.67 | 0.45/0.66 | 0.46/0.68 | 0.39/0.61 |
| Asp–GAC | 0.86/0.56 | 0.84/0.45 | 0.57/0.33 | 0.55/0.34 | 0.54/0.32 | 0.61/0.39 |
| Cys–UGU | 0.43/0.35 | 0.61/0.48 | 0.89/0.61 | 0.91/0.60 | 0.92/0.63 | 0.92/0.58 |
| Cys–UGC | 0.57/0.65 | 0.39/0.52 | 0.11/0.39 | 0.09/0.40 | 0.08/0.37 | 0.08/0.42 |
| Gln–CAA | 0.21/0.30 | 0.56/0.54 | 0.98/0.67 | 0.97/0.66 | 0.97/0.66 | 0.99/0.65 |
| Gln–CAG | 0.79/0.70 | 0.44/0.46 | 0.02/0.33 | 0.03/0.34 | 0.03/0.34 | 0.01/0.35 |
| Glu–GAA | 0.13/0.38 | 0.31/0.54 | 0.95/0.69 | 0.95/0.68 | 0.93/0.68 | 0.96/0.67 |
| Glu–GAG | 0.87/0.62 | 0.69/0.46 | 0.05/0.31 | 0.05/0.32 | 0.07/0.32 | 0.04/0.33 |
| His–CAU | 0.08/0.44 | 0.09/0.54 | 0.28/0.66 | 0.30/0.65 | 0.33/0.67 | 0.27/0.62 |
| His–CAC | 0.92/0.56 | 0.91/0.46 | 0.72/0.34 | 0.70/0.35 | 0.67/0.33 | 0.73/0.38 |
| Amino acid–anticodon | Numbers of tDNA genes | |||||
| Asp–ATC | 0 | 0 | 0 | 0 | 0 | 0 |
| Asp–GTC | 10 | 10 | 16 | 19 | 16 | 16 |
| Cys–ACA | 0 | 0 | 0 | 0 | 0 | 0 |
| Cys–GCA | 3 | 4 | 4 | 3 | 4 | 4 |
| Gln–TTG | 4 | 6 | 9 | 9 | 9 | 9 |
| Gln–CTG | 4 | 4 | 1 | 1 | 1 | 1 |
| Glu–TTC | 3 | 7 | 14 | 15 | 14 | 14 |
| Glu–CTC | 8 | 9 | 2 | 2 | 2 | 2 |
| His–ATG | 0 | 0 | 0 | 0 | 0 | 0 |
| His–GTG | 5 | 4 | 7 | 7 | 7 | 7 |
Favored codons in highly expressed genes relative to less expressed genes are shown in bold (P < 0.05). See Tables 4 and 5, which are published as supporting information on the PNAS web site, for a full list of relative frequencies of codon usage.
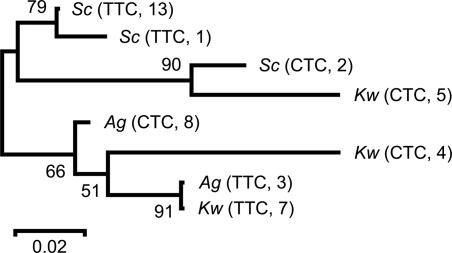
It was proposed that codon-usage bias is generally correlated with overall genome GC content, which is largely determined by mutational processes (15). Moreover, in most prokaryotic genomes, codons that are favored in highly expressed genes are well conserved (16). In our analysis, the codon preferences for these yeast species also agree with their genome GC content, i.e., 44% and 52% for K. waltii and A. gossypii and 38% ≈ 40% for the four Saccharomyces species. However, although most-favored codons are the same among these species (Table 4), we found a switch of the preferred codon of glutamine (Gln) between CAA and CAG and a switch of the preferred codon of glutamic acid (Glu) between GAA and GAG between S. cerevisiae and A. gossypii. As shown in Table 2, these switches might be due to changes in tDNA gene copy number. For instance, the numbers of tDNA-Glu genes for anticodons TTC and CTC are 14 and 2 in S. cerevisiae but 3 and 8 in A. gossypii, and this may explain why the GAA codon is preferred in S. cerevisiae, whereas GAG is preferred in A. gossypii. Such a difference in codon preference can increase the synonymous distance between species. The tDNA gene phylogeny suggests that the change of gene copy number can be derived from a point mutation at anticodon or from duplication/deletion of tDNA genes in the genome (Fig. 3).
Fig. 3.
The neighbor-joining tree of tDNA-Glu genes among three yeast species [S. cerevisiae (Sc), K. waltii (Kw), and A. gossypii (Ag)]. The triplet and number in the parentheses indicate, respectively, the tDNA anticodon and the gene copy number in the corresponding genome. The numbers at branch nodes are bootstrap values. This phylogeny suggests that the switch between anticodons occurred at least twice in the evolution of the tDNA-Glu gene in these yeast species.
Codon-usage bias is a compromise between compositional constraint (genomic GC content) and natural selection acting at the level of translation (17–19). If these two forces act in the same direction, for example, a preferred codon ending in G or C in a GC-rich genome, codon-usage bias could be extremely strong for highly expressed genes. On the other hand, the two forces may counteract each other; for example, a preferred codon ending in G or C in an AT-rich genome may have its frequency only slightly >50% for highly expressed genes. This observation might explain why the high divergence between the K. waltii–A. gossypii and the Saccharomyces sensu stricto species occurred mostly in highly expressed genes.
Gao and Innan (4) estimated the expected length of concerted evolution in S. cerevisiae as 25 million years, based on the theory the same group had proposed earlier (20) (f = 9 of 51; 51 gene pairs shows concerted evolution at the divergence time between S. cerevisiae and S. bayanus, whereas 9 gene pairs are still under concerted evolution at the divergence time between S. cerevisiae and S. paradoxus). We selected 18 gene pairs for which the paralogues and orthologues in S. cerevisiae, S. paradoxus, and S. bayanus are all available and with CAI ≥ 0.7. We detected gene conversion in 11 S. cerevisiae gene pairs. When we used S. paradoxus to calculate the orthologous distance instead, 6 gene pairs still have gene-conversion events detectable. The expected length of concerted evolution for S. cerevisiae genes with CAI ≥ 0.7 thus estimated is 70 million years (f = 6 of 11, from S. cerevisiae–S. bayanus divergence to S. cerevisiae–S. paradoxus divergence). Note that this value may be underestimated because these genes are highly constrained and have evolved slowly. Informative sites indicating gene conversion may be too few to make the statistics significant. However, we obtained a similar estimate by assuming that the duration of concerted evolution started at the WGD event, and the WGD occurred 100 million years ago (f = 12 of 21, from WGD to S. cerevisiae–S. bayanus divergence). Using the same method, we can estimate the expected lengths of concerted evolution for S. cerevisiae genes with CAI between 0.5 and 0.7 and CAI < 0.5 as 20 million years and 10 million years, respectively (f = 4 of 31 and 4 of 238, from WGD to S. cerevisiae–S. bayanus divergence).
In summary, our analysis suggests that codon-usage bias and protein functional conservation might have been more important than gene conversion for the decelerated evolution of WGD duplicate genes in yeasts. Note that gene conversion occurs only occasionally, whereas codon-usage constraint and functional constraint of proteins are constant forces that slow down sequence evolution. Furthermore, the rate of gene conversion decreases as sequence divergence increases. For this reason, gene conversion may not be an effective means for long-term maintenance of sequence similarity between duplicate genes in the absence of codon-usage constraint or functional constraint. In contrast, both codon-usage constraint and protein functional constraint can slow down sequence evolution in the absence of gene conversion. Of course, the three factors can have synergistic effects in maintaining high sequence similarity between paralogues.
Materials and Methods
Sequence Data.
We used the WGD gene pairs in S. cerevisiae and their orthologues in K. waltii (3) and A. gossypii (11) and included their syntenic orthologues from three other species, S. bayanus, S. mikatae, and S. paradoxus (8). All sequences were aligned by using the amino acid sequences with CLUSTAL W 1.83 (21) and back-translated to the DNA level. The KS values were estimated by using PAML 3.14 (22). CAI values (9), each of which indicates the strength of codon-usage bias, were obtained from the Munich Information Center for Protein Sequences (MIPS) (Neuherberg, Germany) (23) for S. cerevisiae genes.
Identification of Gene-Conversion Events.
Numerous methods for gene-conversion identification have been developed, but these methods are either not suitable or not powerful enough for this analysis. For example, S. Sawyer's (24) method uses measures of the distribution of identical synonymous sites between sequence pairs to identify candidate regions of conversion. This method assumes a neutral evolutionary process for synonymous sites and may, therefore, not be suitable for yeast genes in which codon-usage bias affects synonymous substitution. More importantly, this method does not use any outgroup for reference, so it is, in general, less powerful than phylogeny-based methods. Other methods, such as those of Jakobsen and coworkers (25, 26), rely on the examination of site-by-site phylogenies, and the phylogeny for each site in a multiple alignment of paralogues and orthologues is tested for its support of conversion. Although these methods are similar to ours, they suffer when there are multiple substitutions at individual sites (27). Multiple substitutions may, again, be a problem in our analysis, because we are examining the ancient duplicates retained from the WGD in yeast, in which multiple substitutions are common. Therefore, we have developed a related algorithm for conversion identification.
We used WGD orthologues in the four genomes, S. cerevisiae, S. bayanus, S. mikatae, and S. paradoxus. At nucleotide position i, let Di equal the number of nucleotide differences between the two nucleotides in paralogous gene 1 and gene 2 in species 1 (the species under study), and Bji equal the number of nucleotide differences in gene j (j = 1, 2) between species 1 and its orthologue in species 2. Let Bi = (B1i + B2i)/2. Sequences with gaps longer than 50% of the alignment were removed. For a gene under study, species with only one (or no) paralogue available are also removed. Gaps are all removed. For S. cerevisiae, S. paradoxus, or S. mikatae, Bi is calculated between the species under study and S. bayanus. For S. bayanus, Bi is calculated as the average of the differences between S. bayanus and the available three species.
Under the null hypothesis of no gene conversion, the distance (number of differences) between the two paralogues in a species should be larger than or equal to the distance between orthologues, i.e., Di − Bi ≥ 0, because the duplication event occurred before speciation. Dynamic programming is used to select the segment from site m to n that maximizes Σi=mn(Bi − Di). This segment has N sites, where N = n − m + 1. Let D = Σi=mnDi and B = Σi=mnBi. If n ≥ 20, the binomial probability to observe D ≤ B for a segment of N sites is calculated by using the orthologous distance B as the expected distance, i.e., D = B. This is a stringent criterion, because the WGD event occurred earlier than speciation events. The estimated probability is
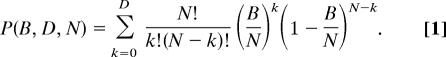 |
However, this segment always has its first and last sites supporting Bi > Di, which may cause an overestimate of the significance. Therefore, we remove the first or the last site of the segment, and recalculate B and D as Σi=m+1nBi and Σi=m+1nDi or Σi=mn−1Bi and Σi=mn−1Di and obtain binomial probabilities P1 and P2, respectively. The higher value of P1 and P2 is used.
The segments thus identified with the paralogous distance significantly smaller than the orthologous distance might potentially be derived from gene conversion. However, many possible segments of N sites can be selected from the entire gene sequence, so we need to take this factor into consideration. Therefore, for each segment with a binomial probability P < 0.01 computed from Eq. 1, we construct an empirical distribution of B for a segment of length N using 10,000 bootstrap samples from {B1, B2, .., BL}, where L equals alignment length for the gene under consideration. Then, it is possible to determine the significance of D by counting the proportion of samples for which D < B. Segments with a binomial probability P < 0.01 and with an empirical probability <0.01 are considered candidate gene conversions.
Codon-Usage Frequencies and tDNA Genes.
Relative frequencies of codon usage in orthologues of WGD genes were calculated for the genomes of K. waltii, A. gossypii, S. cerevisiae, S. bayanus, S. mikatae, and S. paradoxus. Two sets of gene pairs were obtained. S. cerevisiae genes with CAI > 0.5 were classified into the highly expressed set and so were their orthologues in other species, whereas genes with CAI < 0.2 were classified into the less expressed set. The χ2 test was used to examine whether a codon is favored in highly expressed genes compared with less expressed genes. We obtained tDNA genes of S. cerevisiae from the Munich Information Center for Protein Sequences (MIPS), and used the sequences and genomic BLAST in the National Center for Biotechnology Information (NCBI) to identify orthologues in the other five genomes.
Supplementary Material
Acknowledgments
We thank Yu-Ping Poh for discussion and the Structural Bioinformatics Core at the National Chiao Tung University for hardware and software support. This work was supported by National Science Council Grants NSC 094-2917-I-009-015 (to Y.-S.L.) and NSC 093-3112-B-009-001 (to J.-K.H.), the U.S. Department of Education's Graduate Assistance in Areas of National Needs Program (J.K.B.), and National Institutes of Health grants (to W.-H.L.).
Abbreviations
- CAI
codon-adaptation index
- WGD
whole-genome duplication.
Footnotes
The authors declare no conflict of interest.
References
- 1.Petes TD, Hill CW. Annu Rev Genet. 1988;22:147–168. doi: 10.1146/annurev.ge.22.120188.001051. [DOI] [PubMed] [Google Scholar]
- 2.Petes TD. Nat Rev Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 3.Kellis M, Birren BW, Lander ES. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 4.Gao L-Z, Innan H. Science. 2004;306:1367–1370. doi: 10.1126/science.1102033. [DOI] [PubMed] [Google Scholar]
- 5.Coghlan A, Wolfe KH. Yeast. 2000;16:1131–1145. doi: 10.1002/1097-0061(20000915)16:12<1131::AID-YEA609>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 6.Akashi H. Curr Opin Gen Dev. 2001;11:660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 7.Pal C, Papp B, Hurst LD. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 9.Sharp PM, Li W-H. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsh AE, Fraser HB, Wall DP. Mol Biol Evol. 2005;22:174–177. doi: 10.1093/molbev/msh265. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pohlmann R, Luedi P, Choi S, et al. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 12.Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- 13.Tarrio R, Rodriguez-Trelles F, Ayala FJ. Mol Biol Evol. 2001;18:1464–1473. doi: 10.1093/oxfordjournals.molbev.a003932. [DOI] [PubMed] [Google Scholar]
- 14.Christianson ML. Am J Bot. 2005;92:1221–1233. doi: 10.3732/ajb.92.8.1221. [DOI] [PubMed] [Google Scholar]
- 15.Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH. Proc Natl Acad Sci USA. 2004;101:3480–3485. doi: 10.1073/pnas.0307827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rocha EPC. Genome Res. 2004;14:2279–2286. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell JR, Moriyama EN. Proc Natl Acad Sci USA. 1997;94:7784–7790. doi: 10.1073/pnas.94.15.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musto H, Romero H, Zavala A, Jabbari K, Bernardi G. J Mol Evol. 1999;49:27–35. doi: 10.1007/pl00006531. [DOI] [PubMed] [Google Scholar]
- 19.Kliman RM, Irving N, Santiago M. J Mol Evol. 2003;57:98–109. doi: 10.1007/s00239-003-2459-9. [DOI] [PubMed] [Google Scholar]
- 20.Teshima KM, Innan H. Genetics. 2004;166:1553–1560. doi: 10.1534/genetics.166.3.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z. Bioinformatics. 1997;13:555–556. [Google Scholar]
- 23.Mewes HW, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenstern B, Munsterkotter M, Rudd S, Weil B. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawyer S. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- 25.Jakobsen IB, Easteal S. Bioinformatics. 1996;12:291–295. doi: 10.1093/bioinformatics/12.4.291. [DOI] [PubMed] [Google Scholar]
- 26.Jakobsen IB, Wilson SR, Easteal S. Mol Biol Evol. 1997;14:474–484. doi: 10.1093/oxfordjournals.molbev.a025784. [DOI] [PubMed] [Google Scholar]
- 27.Drouin G, Prat F, Ell M, Clarke GDP. Mol Biol Evol. 1999;16:1369–1390. doi: 10.1093/oxfordjournals.molbev.a026047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.