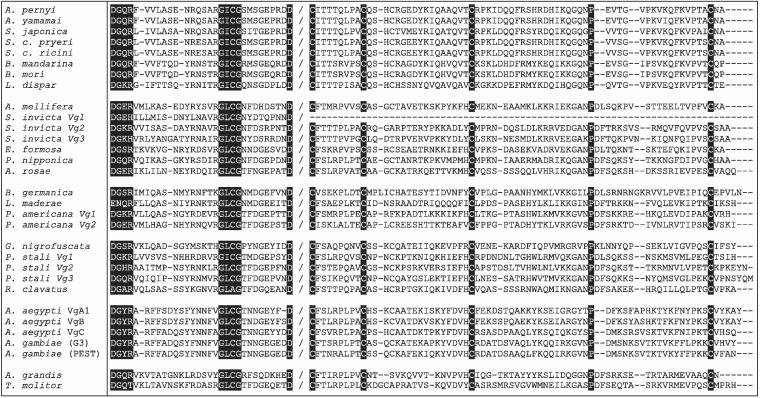
Figure 2.
C-terminal alignment across all 31 available insect Vgs as identified in the legend to Fig. 1. Note that the S. invicta Vg1 sequence appears to be C-terminally truncated. Protein sequences are arranged in six blocks that represent the Lepidoptera, Hymenoptera, Orthoptera, Hemiptera, Diptera and Coleoptera, respectively. The alignment shows the conserved DGXR and GL/ICG motifs and identical aspartic acid (D), cysteine (C) and proline (P) residues in reverse highlight. With one exception, the DGXR and GL/ICG motifs are separated by 17 residues in the Lepidoptera, 19 residues in the Coleoptera and 18 residues in all other insects. For clarity of presentation, only the most proximal and distal parts of the C-terminus are shown, with the sequence break identified by a forward slash. For orientation purposes, the residue immediately preceding the break in T. molitor is D[1660] and that immediately following the break is C[1750].