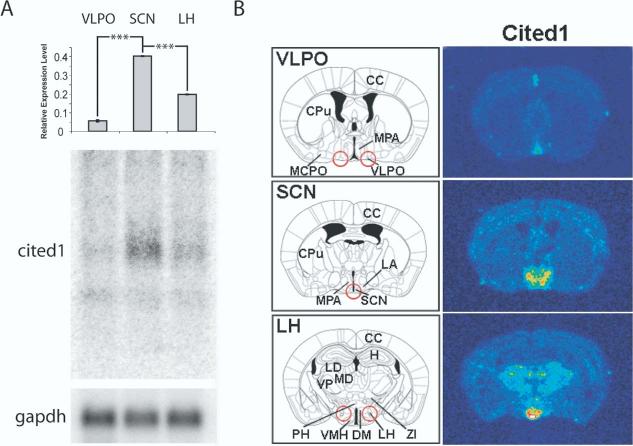
Fig. 1.
Distribution of cited1 mRNA in mouse hypothalamus and forebrain. (A) Northern blots on RNA independent of samples used for microarray analysis identified an enrichment of cited1 mRNA in SCN. The relative expression level of cited1 was normalized to glyceraldehyde phosphate dehydrogenase (gapdh) in each region to control for equal loading. Highest relative expression was observed in SCN, with an increase of 7.4- and 2.1-fold to the VLPO and LH, respectively. Each lane represents RNA pooled from 24 individual animals. Each value represents the average±S.E.M. of three technical replicates. *** P<0.0001 (t-test). (B) Cryostat sections were collected from separate animals at the same level from which tissue punches used for RNA isolation. Red circles indicate punches at the level of VLPO, SCN, or LH. In situ hybridization using 35S-labeled cited1 riboprobe was used to characterize expression patterns. cited1 expression was confined to the MPA including the SCN, dorsohypothalamic area (DM) and VMH and specific nuclei in the thalamus. Schematic diagrams were based on Paxinos and Franklin (2001) and correspond to -0.1, -0.58 and -1.7 mm from Bregma for VLPO, SCN and LH tissue punch regions, respectively. In situ hybridization images were color rendered to enable visualization of labeling (red, highest and blue, lowest levels). CPu, caudate putamen; DM, dorsomedial hypothalamic nucleus; H, hippocampus; MCPO, magnocellular preoptic nucleus; MD, mediodorsal thalamic nucleus; PH, posterior hypothalamic area; ZI, zona incerta.