Abstract
S100P is a member of the S100 family of calcium-binding proteins and there have been several recent reports of its overexpression in pancreatic ductal adenocarcinoma (PDAC). We have used Far Western screening and in vitro interaction assays to identify and confirm a novel target protein for S100P. We have named this protein S100PBPR, and shown that its interaction with S100P is dependent on Ca2+ or Mg2+. S100PBPR was found to localize to cell nuclei where S100P is also present, and the two proteins co-immunoprecipitate. By in situ hybridization, S100PBPR transcript was found in islet cells but not duct cells of the healthy pancreas. Both S100P and S100PBPR were detected by quantitative real-time polymerase chain reaction in pancreatic intraepithelial neoplasia (PanIN) and PDAC samples, and in situ hybridization revealed the presence of S100PBPR transcript in malignant PDAC cells. These data suggest that an interaction between S100P and S100PBPR may be involved in early pancreatic cancer. S100P was further investigated in PanIN lesions and immunohistochemical analysis showed its expression to correlate significantly with increasing grade of PanINs, being found as early as PanIN-1 with more prevalent expression in PanIN-2 and -3. These data suggest that S100P can be added to the genetic progression model for PDAC.
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-related deaths in the industrialized world, with an estimated 30,000 deaths in the United States each year.1 It has a median survival time of 6 months and a 5-year survival of less than 5%, making it one of the most lethal human cancers.2 Except for the recent report of successful use of adjuvant chemotherapy in the ESPAC-1 trail3 there has been no improvement in the survival of pancreatic cancer patients in the last 25 years,1 primarily because of the uniformly advanced stage of disease at time of diagnosis. The ability to recognize and define this malignancy at an early stage is dependent on the identification of novel diagnostic markers indicative of precursor lesions. To date there are no such effective biomarkers.
Precursor ductal lesions have recently been described under the collective term pancreatic intraepithelial neoplasia (PanIN) and are grouped into three histological grades based on increasing degrees of architectural and nuclear atypia.4 PanINs have been examined for loss of heterozygosity at a number of loci and for alterations in a number of genes and proteins that are commonly aberrant in pancreatic carcinomas. These include K-ras, HER-2/neu, p16, p21, p53, DPC4, and BRCA2.5–13 These studies have revealed that PanINs accumulate clonal genetic changes with increasing severity of atypia supporting the theory that they are indeed precursors of ductal adenocarcinomas.14–16
Recent studies using Affymetrix and cDNA microarrays have identified a number of differentially expressed genes in PDAC.17–31 Among these genes S100P has been identified as a potential biomarker for pancreatic adenocarcinoma, being highly up-regulated in pancreatic tumors and cell lines.20,23–26,31,32 S100P is a member of the S100 family of EF-hand, calcium-binding proteins. The S100 family consists of at least 20 members, none of which are ubiquitously expressed.33 They have a variety of functions and target proteins. S100P is one of the least studied members of this family. It is a 95-amino acid protein, first purified from placenta (hence the “P”).34 It has since been detected in gastric, gall bladder, bladder epithelium,32 and in esophageal epithelial cells during differentiation.35 S100P has been associated with cellular immortalization of breast cancer cell lines,36 with doxorubicin resistance in colon cancer cells37 and with androgen independence in prostate cancer.38,39 In addition, we have previously described the up-regulation of S100P in intraductal papillary mucinous neoplasms.40 Decreased survival of patients with lung cancer has also recently been shown to correlate with S100P expression.41 Although the biological effects of S100P remain to be fully elucidated, Arumugam and colleagues42 have demonstrated an interaction between S100P and RAGE that acts in an autocrine manner to stimulate cell proliferation and survival. S100P has also been shown to interact with the membrane/F-actin crosslinking protein ezrin in a calcium-dependent manner43 and is reported to bind to Cacy/SIP, a component of a novel ubiquitination pathway, which leads to degradation of β-catenin.44 In this study we have isolated a novel binding partner for S100P, S100PBPR (S100P binding protein Riken), and analyzed the expression of both these proteins in PanIN lesions.
Materials and Methods
Tissues and Cell Lines
Frozen and paraffin-embedded pancreatic tissues were obtained from the Human Biomaterials Resource Centre, Department of Histopathology, Charing Cross Hospital, London, UK, and the Department of Pathology, University of Kiel, Kiel, Germany, with full ethical approval of the host institutions. PanIN specimens were obtained from patients under the care of Dr. Teresa A. Brentnall, University of Washington, Seattle, WA. A panel of 10 pancreatic cancer cell lines was used in this study: FA6, IMIMPC2, Mia-Paca2, PT45, Paca44, Panc1, HPAF, MDA Panc3, SUIT2, and T3M4. All cancer cell lines were obtained from the Cancer Research UK Cell Services (Clare Hall, Middlesex, UK). These cell lines were cultured in E4 medium (Cancer Research UK Media Production, Clare Hall, Middlesex, UK) supplemented with 10% heat-inactivated fetal calf serum (GibcoBRL, Life Technologies, Paisley, UK). HeLa and 293 cells were also used in this study and were cultured as described above. The human pancreatic duct epithelial line, HPDE, was a kind gift from Dr. Ming-Sound Tsao, University of Toronto, Toronto, Canada, and was grown in keratinocyte growth medium (GibcoBRL, Life Technologies) as described by Furukawa and colleagues.45
Plasmids
IMAGE clone 3448490 encoding full-length S100P was obtained from Mammalian Gene Collection (MRC Gene Service, Babraham, Cambridgeshire, UK). S100P was amplified from IMAGE clone 3448490 using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) and subcloned into vector pGEX-2TK (Amersham Biosciences UK Ltd., Bucks, UK) in-frame with the GST-tag to create the plasmid pGEX-S100P. The primer sequences used for pGEX-S100P were: forward 5′-ATCAGGGATCCATGACGGAACTAGAGACAGCCA-3′ and reverse 5′-ACGATGAATTCTCATTTGAGTCCTGCCTTCTCA-3′(restriction endonuclease recognition sites for BamHI and EcoRI are underlined). Using the pcDNA3.1/V5-His TOPO TA expression kit (Invitrogen Life Technologies, Carlsbad, CA), S100P was also subcloned into vector pcDNA3.1/V5-His in-frame with the V5-His-tag, to create plasmid pcDNA3.1-S100P-V5-His. The primer sequences used for pcDNA3.1-S100P-V5-His were: forward 5′-ATGACGGAACTAGAGACAGCCA-3′ and reverse 5′-TTTGAGTCCTGCCTTCTCA-3′.
The full coding sequence of S100PBPR was obtained from universal cDNA by polymerase chain reaction (PCR) with Pfu Turbo DNA polymerase (Stratagene). Full-length S100PBPR was cloned into pcDNA3.1+ vector (Invitrogen Life Technologies) creating pcDNA3.1-S100PBPR. The primer sequences used for pcDNA3.1-S100PBPR were: forward 5′-ATCAGGGATCCATGATGTGCTCAC-GGGTGCC-3′ and reverse 5′-ACGATGAATTCTTAACTGTACGAGAAGTCTGGGA-3′ (restriction endonuclease recognition sites for BamHI and EcoRI are underlined). Full-length S100PBPR was also cloned in-frame with the green fluorescent protein (GFP) of vector pEGFP-C2 (BD Biosciences Clontech, Palo Alto, CA) creating plasmid pEGFP-S100PBPR. The primer sequences used for pEGFP-S100PBPR were: forward 5′-ATCAGGAATTCATGATGTGCTCACGGGTGCC-3′ and reverse 5′-ACGATGGATCC-TTAACTGTACGAGAAGTCTGGGA-3′ (restriction endonuclease recognition sites for BamHI and EcoRI are underlined). Finally S100PBPR was cloned into vector pCMV-Tag2B (Stratagene) in-frame with the FLAG-tag, creating vector pCMV-Tag-S100PBPR. The primer sequences used were: forward 5′-ATCAGGGATCCATGATGTGCTCACGGGTGCC-3′ and reverse 5′-ACGATGAATTCTTAACTGTACGAGAAGTCTGGGA-3′ (restriction endonuclease recognition sites for BamHI and EcoRI are underlined). All constructs were verified by sequence analysis.
Far Western Screening Assay
The S100P probe for Far Western (FW) screening was produced by in vitro translation of a full-length S100P cDNA IMAGE clone 3448490 using the TNT coupled reticulocyte lysate system (Promega Biosciences, Madison, WI). A luciferase plasmid was used as a control for efficiency of in vitro translation and later as a negative control for the FW assay. The proteins were radiolabeled in 100-μl reactions using l-[35S]methionine/l-[35S]cysteine Promix (Amersham Biosciences UK Ltd.). Unincorporated amino acids were removed using Microcon-MWCO 3000 filters (Millipore Corp., Bedford, MA), according to the manufacturer’s instructions and the labeled product resuspended in FW buffer (25 mmol/L Tris-HCl, pH 7.5, 100 mmol/L KCl, 5 mmol/L MgCl2, 0.1% Tween 20, 5% glycerol, 1 mmol/L dithiothreitol) with protease inhibitors (Complete EDTA-free; Roche Diagnostics GmbH, Mannheim, Germany). Successful synthesis and purification of the probe was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. A placental λ TriplEx phage library (BD Biosciences Clontech), comprising 6.4 × 105 clones, was screened. The Escherichia coli strain XL1-Blue (Stratagene) was transduced with the λ TriplEx phage library and plated at ∼100 plaque-forming units per 10 cm Petri dish, according to the λ TriplEx library manual (Clontech PT3003-1). Plates were incubated at 42°C for 3 to 4 hours until plaques were just visible, then overlaid with Hybond-C Extra membrane (Amersham Biosciences UK Ltd.), soaked in 10 mmol/L isopropyl-β-d-thiogalactopyranoside, and incubated at 37°C for a further 4 hours. The membranes were then removed from the Petri dishes and placed in blocking solution (FW buffer containing 5% dry milk) for 2 hours, shaking gently at room temperature. After blocking, the membranes were placed in FW buffer, supplemented with in vitro translated, 35S-labeled protein probe and incubated overnight at 4°C, with gentle shaking. The membranes were washed several times in FW buffer; each wash was performed for 30 minutes at room temperature with gentle shaking. Washed membranes were air-dried and exposed to BioMax MR-1 (Eastman Kodak Co., Rochester, NY) film for 5 to 7 days.
Expression of S100P-GST in E. coli
Plasmids pGEX-S100P and vector alone, pGEX-2TK, were introduced into the E. coli strain pLysS (Promega Biosciences). The bacteria were cultured in LB medium with ampicillin to an OD 600 of 0.4 to 0.6 at 37°C. Protein expression was induced with 1 mmol/L IPTG and bacteria were cultured for a further 4 hours at room temperature to enhance correct protein folding. Cells were pelleted and protein lysates made by repeat freeze thawing and sonication in lysis buffer (20 mmol/L Tris-HCl, 300 mmol/L NaCl, 10% glycerol, 0.5% Nonidet P-40, 2 mmol/L dithiothreitol, 1 mg/ml lysozyme) with protease inhibitors (Complete EDTA-free, Roche Diagnostics GmbH) at 4°C.
In Vitro Binding Assays
Plasmid pcDNA3.1-S100PBPR was in vitro translated and radiolabeled, as described for FW probes. S100P-GST or GST alone, immobilized on glutathione agarose, was incubated with radiolabeled target protein at 4°C, overnight, in CRB300 buffer (20 mmol/L Tris-HCl, 300 mmol/L NaCl, 10% glycerol, 0.5% Nonidet P-40, 2 mmol/L dithiothreitol) with protease inhibitors (Complete EDTA-free, Roche Diagnostics GmbH). Glutathione agarose was washed three times in CRB300 buffer followed by three washes in CRB100 buffer (CRB300 buffer with 100 mmol/L NaCl). To determine the dependence of the interaction between S100P and S100PBPR on the presence of divalent cations, buffers CRB300 and CRB100 were supplemented with calcium chloride, magnesium chloride, or 1 mmol/L ethylenediaminetetraacetic acid (EDTA). Calcium chloride was added at concentrations ranging from 10 pmol/L to 5 mmol/L. Magnesium chloride was added at concentrations ranging from 100 pmol/L to 100 mmol/L. Bound target proteins were eluted in 2× SDS-PAGE sample buffer (90 mmol/L Tris-HCl, 20% glycerol, 2% SDS, 0.02% bromophenol blue, 100 mmol/L dithiothreitol) at 100°C, separated by SDS-PAGE on 10% polyacrylamide resolving gels, and detected by autoradiography.
Cell Transfection and Immunofluorescence Microscopy
HeLa cells grown to 60% confluency in chamber slides (Lab-Tek; Nalge Nunc Int., Naperville, IL) were transiently transfected with plasmid pEGFP-S100PBPR encoding the fusion protein S100PBPR-GFP, or with pEGFP-C2 vector encoding GFP alone. FuGENE 6 transfection reagent (Roche Diagnostics GmbH) was used according to the manufacturer’s protocol. Cells were cultured for a further 24 hours then fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 minutes at room temperature and washed three times with PBS. Cells were permeabilized with 0.1% Triton X-100 in PBS for 5 minutes at 4°C, followed by three washes in PBS. Endogenous S100P was detected by incubating the cells with an anti-human S100P monoclonal antibody (BD Biosciences Pharmingen) at a 1:25 dilution in PBS for 30 minutes at room temperature. After washing three times in PBS, the primary antibodies were detected with Alexa Fluor 568 goat anti-mouse secondary antibodies (Molecular Probes, Leiden, The Netherlands) at a dilution of 1:1000 in PBS. Cells were washed three times and the DNA stained with 4,6-diamidino-2-phenylindole (Molecular Probes) at a dilution of 20 μg/ml in PBS. After three further washes in PBS, slides were mounted using Permafluor aqueous mounting medium (Immunotech, Marseille, France) and viewed using an Olympus BX51 fluorescent microscope.
Co-Immunoprecipitation of S100P and S100PBPR
293 cells grown to 60% confluency were transiently co-transfected with plasmids pCMV-Tag-S100PBPR and pcDNA3.1-S100P-V5-His using FuGENE 6 transfection reagent (Roche Diagnostics GmbH), according to the manufacturer’s protocol. Cells to be used as negative controls for co-immunoprecipitation assays were co-transfected with either pCMV-Tag-S100PBPR or pcDNA3.1-S100P-V5-His and an empty vector. Cells were lysed 48 hours after transfection with reporter lysis buffer (Promega), according to the manufacturer’s instructions. Protein extracts were quantitated using Bio-Rad protein assay (Bio-Rad Laboratories, Richmond, CA).
Five hundred μg of total protein were used per co-immunoprecipitation reaction. Immunoprecipitation of S100PBPR-FLAG was conducted using anti-FLAG M2 affinity gel (Sigma-Aldrich, St. Louis, MO), following the manufacturer’s protocol. Bound proteins were eluted using 2× SDS sample buffer and were separated on 15% polyacrylamide gels. Proteins were transferred onto nitrocellulose membrane and subjected to standard Western blotting analysis. S100P-V5-His was detected using anti-V5 antibody (Invitrogen Life Technologies) at a dilution of 1:5000. Primary antibodies were detected using the anti-mouse horseradish peroxidase-conjugated secondary antibodies (1:1000) (Autogen Bioclear, Wiltshire, UK) and visualized using ECL Western blotting detection reagents (Amersham Biosciences, Buckinghamshire, UK). Before co-immunoprecipitation experiments, the level of S100P-V5-His protein expression was determined by Western blot as outlined above. Twenty μg of total protein was used for each cell lysate analyzed.
Reverse Transcriptase (RT)-PCR
RNA samples from normal pancreas, brain, breast, spleen, liver, and lung were obtained from Ambion Inc. (Austin, TX). cDNAs were synthesized from 1 μg of total RNA using an oligo dT primer and the Multiscribe reverse transcription kit (Applied Biosystems, Warrington, Cheshire, UK) as instructed by the manufacturer. Reverse transcription was followed by 30 PCR cycles (1 minute of denaturation at 94°C, 1 minute of annealing at 55°C, and 1 minute of extension at 72°C). Primers designed for human S100PBPR are as follows: forward 5′-TCCTTGCTGGAGCTGTCAGAG-3′ and reverse 5′-ACTGGTCCCAGGCTGTACGA-3′. Primers for 18S ribosomal RNA, which was used as an endogenous control, are as follows: forward 5′-CGCCGCTAGAGGTGAAATTC-3′ and reverse 5′-CATTCTTGGCAAATGCTTTCG-3′. Amplified products were separated on 1.5% agarose gels and visualized with ethidium bromide.
Quantitative Real-Time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen Life Technologies) according to the manufacturer’s protocol. PDAC samples were enriched for a tumor cell content of 60 to 80%, before RNA extraction, by a process of block trimming guided by frequent hematoxylin and eosin (H&E) control sections, as previously described.40 cDNAs were synthesized from 1 μg of total RNA using random hexamers and the Multiscribe reverse transcription kit (Applied Biosystems) as instructed by the manufacturer.
PCR reactions containing 10 ng of cDNA, SYBR Green sequence detection reagents (Applied Biosystems), and gene-specific primers were assayed on an ABI7700 sequence detection system (Applied Biosystems). Primers designed for human S100P are as follows: forward 5′-TGCAGAGTGGAAAAGACAAGGAT-3′ and reverse 5′-CCACCTGGGCATCTCCATT-3′. Primers designed for human S100PBPR are as follows: forward 5′-CCAGGACTAATGTTCCGACGTT-3′ and reverse 5′-CTGGGTCTTCTATATGAGCAATGACA-3′. These primers amplify a 75-bp S100P product and a 93-bp product for S100PBPR. The accumulation of PCR products was measured in real time as the increase in SYBR-Green fluorescence. Data were analyzed using the Sequence Detector program v1.9.1 (Applied Biosystems). All reactions were performed in triplicate. Quantification of gene expression was conducted as described by the manufacturer using 18S mRNA as an internal standard. Gene expression patterns in PanIN, tumor, and chronic pancreatitis (CP) samples were compared to the value of normal donor pancreas, arbitrarily set at 1. Quantitative gene expression levels of pancreatic cancer cell lines were compared to the value for primary cells derived from normal ducts (a kind gift of Dr. T. Hollingsworth, Eppley Institute, Omaha, NE).
In Situ Hybridization
The DNA sequence for the S100PBPR probe for in situ hybridization was amplified by PCR from normal spleen cDNA using Taq polymerase (Roche Diagnostics GmbH). The primers used amplify a 212-bp product, corresponding to S100PBPR and are as follows: forward 5′-TCCTTGCTGGAGCTGTCAGAG-3′ and reverse 5′-ACTGGTCCCAGGCTGTACGA-3′. This product was cloned into the vector pCR4-TOPO using the TOPO cloning kit (Invitrogen Life Sciences) to create pCR4-S100PBPR-ISH. Positive clones were verified by sequence analysis.
pCR4-S100PBPR-ISH DNA was linearized with NotI for anti-sense probe and PstI for sense probe. Restriction endonucleases were removed by phenol:chloroform:isoamyl alcohol extraction and the linear template DNA was precipitated. Riboprobes were synthesized from 1 μg of template DNA and digoxigenin (DIG) labeled using a DIG RNA labeling kit (Roche Diagnostics GmbH), according to the manufacturer’s instructions. T3 and T7 polymerases were used to synthesize anti-sense and sense probes, respectively. DIG-labeled riboprobes were precipitated and resuspended in diethyl pyrocarbonate-treated water. DIG incorporation was verified using DOT blot with anti-digoxigenin-AP Fab fragments (Roche Diagnostics GmbH) diluted 1:5000.
Anti-sense and sense riboprobes for S100PBPR were hybridized to freshly cut 5-μm-thin sections of formalin-fixed, paraffin-embedded pancreatic tissue. Three hundred ng of DIG-labeled probe were used per slide. Sense probe was used as a negative control. In situ hybridization was performed on sections of healthy pancreas and five different PDAC specimens using the Ventana Discovery System with Ventana Ribomap and Bluemap kits as instructed by the manufacturer (Ventana Medical Systems, Tucson, AZ).
Immunohistochemistry
Paraffin-embedded tissues from 41 resection specimens were cut into 5-μm-thin sections and stained with H&E. Blocks were selected on the basis of the content of PDAC or PanIN lesions. PanINs were classified according to previously established criteria.46 The selected cases were immunostained with anti-S100P antibody. After antigen demasking for 2 minutes with the pressure cooker method, 5-μm-thin sections were routinely deparaffinized and blocked with normal rabbit serum (1:20 diluted in PBS) for 30 minutes. Primary S100P antibody (1:400 dilution; BD Biosciences Pharmingen) was added for 1 hour. After washing (2 × 5 minutes) in PBS the slides were incubated with the secondary antibody for 30 minutes, then washed again in PBS (2 × 5 minutes), followed incubation with 0.3% hydrogen peroxide in methanol for 25 minutes. Streptavidin (1:300 in PBS) was added for 30 minutes and after washing with PBS (2 × 5 minutes) the reaction was visualized by diaminobenzidine (DAB) reaction using a chromogen tablet system (DAKO, Hamburg, Germany).
Statistical evaluation was performed using a two-sided Fisher’s exact test, using R on a Unix platform. A P value <0.05 was accepted as statistically significant. Levels of expression between PanIN-1A and -1B, PanIN-1B and PanIN-2, PanIN-2 and PanIN-3, and PanIN-3 and PDAC lesions were compared.
Results
S100P Binds a Novel Target Protein, S100PBPR
Far Western (FW) screening was used to isolate S100P target proteins. This technique was favored over yeast two-hybrid screening because the calcium requirement for target protein binding by S100 proteins is probably not met in the environment of the yeast nucleus.47 In total 6.4 × 105 clones from a placental λ TriplEx library were screened, resulting in 14 positive clones after primary screening. After second and third round screening six clones were found to be false positives, whereas eight clones were consistently found to interact with S100P. On sequencing and extensive BLAST database searches four of these clones had no significant similarity to any previously characterized sequence, whereas the remaining four displayed homology to anonymous cDNAs represented in the data base. Of these the largest clone corresponded to the amino-terminal 969 bp of a RIKEN cDNA, BC015175 (NCBI accession number) also listed as hypothetical protein FLJ12903 (Hs.369253). As we have confirmed the interaction between the protein encoded by cDNA BC015175 and S100P, we have named this protein S100PBPR (binding protein Riken) and it will be referred to as such throughout the rest of this study.
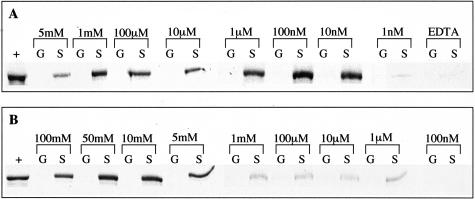
S100PBPR is situated at 1p34.3 and has a 1224-bp coding region, which encodes a 408-amino acid protein. It shares no sequence or structural similarity to any known protein at present. The interaction between S100P and S100PBPR was confirmed using in vitro binding assays using S100P-GST fusion protein. It has previously been demonstrated that the EF-hand motifs of S100P bind Ca2+ and Mg2+ at concentrations within the physiological ranges of these ions.48 To assess the dependence of the S100P/S100PBPR interaction on divalent cations, in vitro binding assays were conducted in buffer containing Ca2+ (5 mmol/L to 10 pmol/L), Mg2+ (100 mmol/L to 100 pmol/L), or 1 mmol/L EDTA. Figure 1A shows the results for in vitro binding assays between S100P-GST (labeled “S”) or GST alone (labeled “G”) and 35S-S100PBPR in the presence of Ca2+ at concentrations decreasing from 5 mmol/L to 1 nmol/L or in the presence of 1 mmol/L EDTA. The proteins were found to interact at Ca2+ concentrations between 5 mmol/L and 10 nmol/L, with hardly any visible interaction at a Ca2+ concentration of 1 nmol/L (Figure 1A) and none less than 1 nmol/L Ca2+. The interaction between S100P and S100PBPR was completely abolished in buffer containing EDTA (Figure 1A). There was no visible interaction between GST alone and S100PBPR under any of the experimental conditions. Figure 1B shows the results of in vitro binding assays conducted in the presence of Mg2+ at concentrations decreasing from 100 mmol/L to 100 nmol/L. S100P-GST and S100PBPR were found to interact at Mg2+ concentrations between 100 mmol/L and 1 μmol/L but not at a Mg2+ concentration of 100 nmol/L. There was no interaction between GST alone and S100PBPR in the presence of Mg2+.
Figure 1.
S100P and S100PBPR interact in the presence of Ca2+ or Mg2+. GST alone (columns G) or S100P-GST (columns S) immobilized on glutathione agarose were incubated overnight at 4°C with 35S-labeled S100PBPR. Incubations and washes were conducted in the presence of Ca2+, 1 mmol/L EDTA, or Mg2+. A: The results of in vitro binding assays conducted in the presence of Ca2+ at concentrations ranging from 5 mmol/L to 1 nmol/L or in the presence of 1 mmol/L EDTA. B: The results of in vitro binding assays conducted in the presence of Mg2+ at concentrations ranging from 100 mmol/L to 100 nmol/L. Concentrations of Ca2+, Mg2+, or EDTA are indicated above the relevant lanes. Lanes labeled “+” contain 35S-S100PBPR alone, used as a positive control, detected as a band of ∼60 kd. Lanes labeled “G” contain GST-only samples, which were incubated with 35S-S100PBPR to provide a negative control. Lanes labeled “S” contain S100P-GST samples incubated with 35S-S100PBPR; the presence of a band in these lanes shows an interaction between S100P and S100PBPR at the indicated concentrations of Ca2+ or Mg2+. In A S100P-GST can be seen to interact with S100PBPR at Ca2+ concentrations between 5 mmol/L and 10 nmol/L, the proteins do not interact at Ca2+ concentrations of 1 nmol/L or less or in the presence of 1 mmol/L EDTA. In B S100P-GST can be seen to interact with S100P-BPR at Mg2+ concentrations between 100 mmol/L and 1 μmol/L, the proteins do not interact at Mg2+ concentrations of 100 nmol/L or less. GST alone does not interact with S100PBPR.
Cellular Localization of S100P and S100PBPR
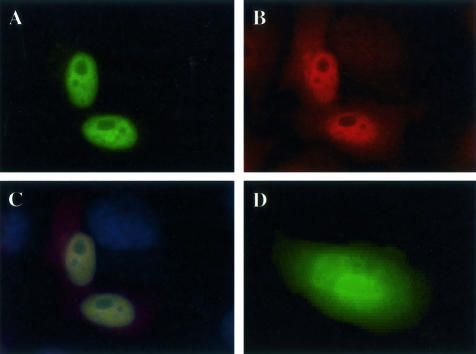
As the cDNA encoding S100PBPR was a result of a large-scale sequencing project, no reagents are currently available to enable its immunological detection. Therefore, we used GFP-tagged S100PBPR transfected into HeLa cells to determine its subcellular localization. S100PBPR-GFP was found to localize to the nucleus (Figure 2A), whereas GFP alone was present throughout the cell (Figure 2D). Immunocytochemical analysis of endogenous S100P in HeLa cells revealed both nuclear and cytoplasmic localization (Figure 2B) suggesting that interaction of S100P and S100PBPR probably occurs in the nucleus.
Figure 2.
Cellular localization of S100P and S100PBPR. HeLa cells were transiently transfected with pEGFP-S100PBPR or the pEGFP vector and endogenous S100P was detected by immunocytochemistry using S100P monoclonal antibodies and Alexa Fluor 568 goat anti-mouse secondary antibodies. A: Nuclear expression of S100PBPR-GFP in transfected HeLa cells. B: Endogenous expression of S100P protein in HeLa cells. C shows both S100PBPR-GFP and endogenous S100P and D shows cells expressing GFP alone. Cells were observed using a fluorescence microscope. S100PBPR-GFP (A) localizes to the nucleus unlike GFP alone (D), which is found throughout the cell. S100P localizes with S100PBPR-GFP in the nucleus (C) but can also be found in the cytoplasm (B).
Co-Immunopreciptiation of S100P and S100PBPR
Because there are no antibodies available for the detection of S100PBPR, to demonstrate an in vivo interaction between S100P and S100PBPR we used exogenous expression of S100P-V5-His and S100PBPR-FLAG in 293 cells. The expression levels of S100P-V5-His in control and test cell lysates were determined by Western blot (Figure 3A). S100PBPR was immunoprecipitated with anti-FLAG M2 affinity agarose and Western blot confirmed the presence of S100P-V5-His. In this system S100P was found to co-immunoprecipitate with S100PBPR (Figure 3B).
Figure 3.
Co-immunoprecipitation of S100P and S100PBPR.293 cells were transiently co-transfected with expression vectors encoding S100P-V5-His and S100PBPR-FLAG. In both A and B, the samples in lanes 1 to 3 consist of 293 cells transfected with S100PBPR-FLAG expression vector and pcDNA3.1/V5-His vector only (lane 1), expression vector for S100P-V5-His and pCMV-Tag vector only (lane 2), S100PBPR-FLAG and S100P-V5-His expression vectors (lane 3). Samples in lanes 1 and 2 acted as negative controls in co-immunoprecipitation experiments. A: Standard Western blot to determine the expression levels of S100P-V5-His in transfected cell lysates. S100P-V5-His was detected with an anti-V5 antibody (Invitrogen Life Technologies) as a band just under 15 kd. Twenty μg of total protein was loaded for each sample in A. From A it can be seen that the samples in lanes 2 and 3 express approximately equal amounts of S100P-V5-His. B: Co-immunoprecipitation experiment. S100PBPR-FLAG was immunoprecipitated using anti-FLAG M2 affinity gel (Sigma-Aldrich). Samples were eluted in 2× SDS-sample buffer and submitted to standard Western blotting techniques for S100P-V5-His, as outlined above. The presence of a band for S100P-V5-His in lane 3 of B indicates S100P does co-immunoprecipitate with S100PBPR. This band was not present in lanes 1 and 2, which were negative controls, as detailed above.
S100PBPR Expression in Normal Tissues
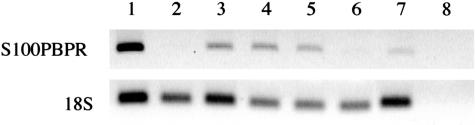
RNAs from normal pancreas, brain, breast, spleen, liver, and lung tissues were screened for S100PBPR expression by RT-PCR. S100PBPR was found to be present in normal brain, breast, spleen, and lung, and virtually absent in normal pancreas and liver (Figure 4).
Figure 4.
S100PBPR expression in normal tissues. cDNAs made from RNA samples isolated from normal organs were used as templates for RT-PCR to determine the presence of S100PBPR expression. Samples were run in the following order: lane 1, universal RNA (positive control); lane 2, pancreas; lane 3, brain; lane 4, breast; lane 5, spleen; lane 6, liver; lane 7, lung; lane 8, negative control. S100PBPR expression was found to be present in brain, breast, spleen, and lung, while virtually absent in normal pancreas and liver.
Expression of S100P and S100PBPR in Pancreatic Cancer
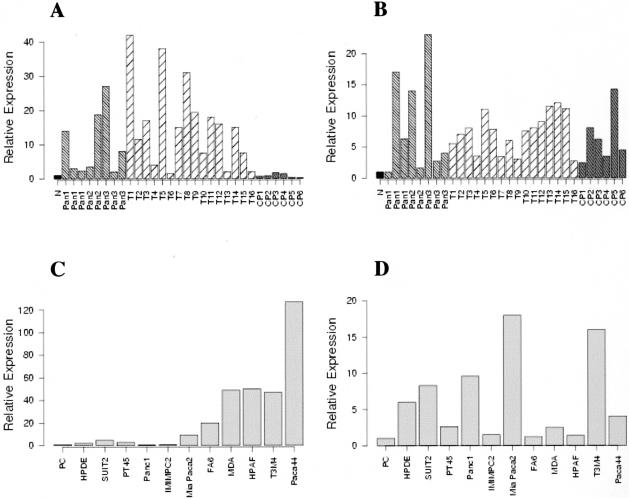
Overexpression of S100P has previously been reported in PDAC.20,23–26,31,32 However, the stage of pancreatic cancer at which S100P is expressed is at present unknown. To deduce this and the status of S100PBPR in pancreatic cancer, quantitative real-time PCR for S100P and its binding partner was conducted on 8 PanIN samples, 16 PDAC specimens, 6 cases of CP, and 10 pancreatic cancer cell lines (Figure 5; A to D).
Figure 5.
Expression of S100P and S100PBPR in PanINs, PDACs, CPs, and pancreatic cancer cell lines, as determined by quantitative real-time PCR. Quantitative real-time PCR was conducted on 10-ng cDNA samples using the SYBR Green method and the ABI7700 sequence detection system. All reactions were conducted in triplicate and 18S mRNA was used as an internal standard. Values for the controls, normal donor pancreas (N) (A and B), and primary cells isolated from normal ducts (PC) (C and D) have arbitrarily been set at 1. A and B show expression levels of S100P and S100PBPR, respectively, in PanINs (Pan), PDACs (T1-16), and CPs (CP1-6), compared to normal donor pancreas. C and D show expression levels of S100P and S100PBPR in pancreatic cancer cell lines compared to primary cells. In each figure the relative expression level is indicated on the y axis as a fold-change compared to normal controls.
Figure 5A shows S100P expression in PanIN, PDAC, and CP samples. S100P was found to be up-regulated more than fourfold in 4 of 8 PanIN samples and in 13 of 16 PDACs, whereas there was no up-regulation of S100P transcript in any of the CP samples. Figure 5B shows the results for S100PBPR expression levels in PanIN, PDAC, and CP samples. As for S100P, S100PBPR was also up-regulated in PanIN and PDAC cases, with greater than fourfold up-regulation in 5 of 8 PanIN samples and 12 of 16 PDAC specimens. Four of six CP samples also revealed greater than fourfold up-regulation of S100PBPR. Figure 5, C and D, shows expression levels of S100P and S100PBPR, respectively, in pancreatic cell lines compared to primary ductal cells. S100P was up-regulated greater than fourfold in Mia Paca2, MDA Panc3, FA6, SUIT2, HPAF, T3M4, and Paca44 cell lines, and interestingly more than twofold in the immortalized cell line HPDE, which was derived from normal duct cells. S100PBPR was up-regulated greater than fourfold in Mia Paca2, Panc1, T3M4, and Paca44, as well as in HPDE.
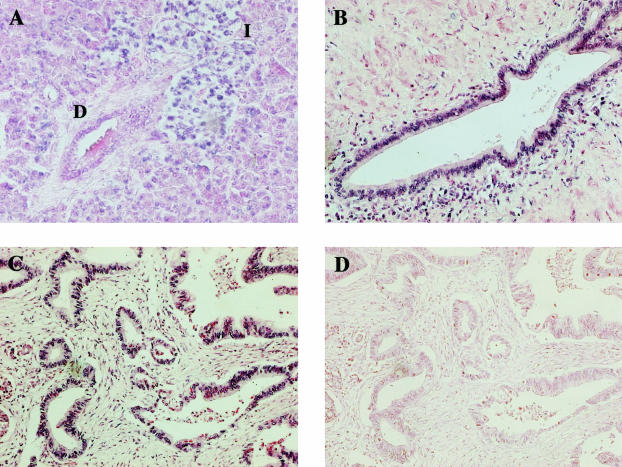
To determine which cells express S100PBPR in both normal pancreas and PDAC specimens we conducted in situ hybridization experiments. Results for S100PBPR in situ hybridization on a section of normal pancreas are shown in Figure 6A. Interestingly, in the normal pancreas, S100PBPR mRNA was detected in the islet cells and at a low level in a proportion of the acinar cell population but not in normal ductal cells (Figure 6A). Figure 6, B and C, shows representative S100PBPR in situ hybridization results from two different PDAC samples. In each of the five PDAC cases studied, S100PBPR mRNA was found in the malignant PDAC cells, in addition to the islet cells and at varying levels in the acinar cell population. S100PBPR transcript was also found in the infiltrating lymphocyte population present in some PDAC samples. A sense probe for S100PBPR was used as a negative control (Figure 6D).
Figure 6.
S100PBPR expression in normal pancreas and PDACs, as determined by in situ hybridization. In situ hybridization was conducted using the Ventana Discovery System with digoxigenin-labeled riboprobes. The presence of S100PBPR mRNA is indicated by a blue signal. A: S100PBPR expression in normal pancreas. B and C: S100PBPR expression in two representative PDAC samples. D: In situ hybridization conducted with a sense S100PBPR riboprobe, used as a negative control. S100PBPR was expressed in the islets of normal pancreas (I) and at a much lower level in the acinar cell population. It was not present in the ductal cells (D) of normal pancreas (A). However, in PDAC S100PBPR mRNA was detected in the malignant ductal epithelial cells (B and C). Original magnifications, ×200.
Unlike S100PBPR, a specific antibody is available for S100P; therefore, to confirm the presence of S100P protein in PanINs we performed immunohistochemical analysis on 41 cases of PDAC with associated PanIN lesions. A PanIN was considered to be positive for S100P if more than 10% of cells within the lesion expressed S100P. The 41 cancer cases provided a total of 227 PanIN lesions for analysis; these comprised 126 PanIN-1A, 52 PanIN-1B, 32 PanIN-2, and 17 PanIN-3 lesions (Table 1).
Table 1.
Results of S100P Immunohistochemistry in PDAC and Associated PanINs
| No. of lesions/cases | No. of S100P lesions/cases | Percentage | |
|---|---|---|---|
| PanIN-1A | 126 | 3 | 2 |
| PanIN-1B | 52 | 7 | 13 |
| PanIN-2 | 32 | 10 | 31 |
| PanIN-3 | 17 | 7 | 41 |
| PDAC | 41 | 38 | 92 |
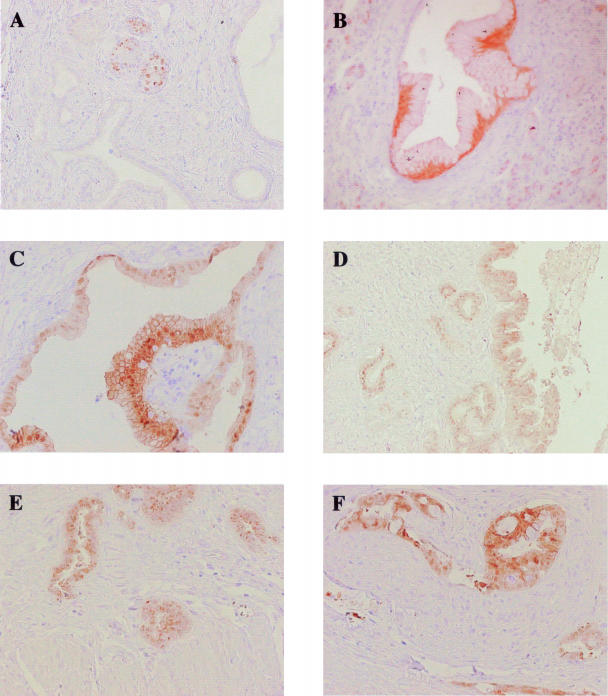
Two percent of PanIN-1A (3 of 126) and 13% of PanIN-1B (7 of 52) expressed S100P. Figure 7B shows a representative image of S100P protein expression in PanIN-1B. Thirty-one percent of PanIN-2 lesions (10 of 32) were positive for S100P (Figure 7C). The progressive increase in the proportion of S100P expressing lesions from PanIN-1A to PanIN-1B and PanIN-1B to PanIN-2 was found to be statistically significant (P < 0.05). The proportion of lesions that expressed S100P also increased to 41% for PanIN-3 (7 of 17) (Figure 7D). Ninety-two percent of invasive PDAC cases (38 of 41) expressed S100P (Figure 7, E and F); this was found to be a significant increase from the proportion of PanIN-2 and -3 lesions positive for S100P (P < 0.01).
Figure 7.
Immunohistochemical analysis of S100P in normal pancreas, PanIN lesions, and PDAC. Representative images of S100P immunoreactivity in normal pancreatic islets (A), PanIN-1B (B), PanIN-2 (C), PanIN-3 (D), PDAC extending into the muscle wall (E), and perineural invasion (F). Original magnifications: ×100 (A, D, E); ×200 (B, C, F).
Normal ducts showed no expression of S100P, whereas nuclear staining was apparent in some islet cells, these cells appeared to be randomly distributed throughout the islet (Figure 7A). Scattered centroacinar cells exhibited variable amounts of cytoplasmic immunoreactivity as well. The S100P staining pattern in PDACs was both cytoplasmic and nuclear, as already described;32 24 cases showed granular cytoplasmic staining that was more intensely perinuclear in 13 samples, 9 of the 41 cases exhibited predominantly nuclear staining, whereas in 7 cases staining was both nuclear and cytoplasmic.
Discussion
In this study we have identified and confirmed an interaction between S100P and a novel protein, S100PBPR. We have shown S100PBPR to be a binding partner of S100P by in vitro binding assays and in vivo, by co-immunoprecipitation from transfected cell lysates. In vitro, we found the interaction between these proteins to be dependent on the presence of divalent cations, Ca2+ and Mg2+. This is consistent with a study by Gribenko and Makhatadze,48 which found that the EF-hands of S100P are able to bind Ca2+ and Mg2+, but not Zn2+, when these ions are at physiological concentrations. It also revealed that metal binding was accompanied by conformational changes in S100P that result in the exposure of hydrophobic surfaces. In a subsequent study, the same authors showed that binding of a target peptide to Ca2+-S100P was driven by hydrophobic interactions.49
Very little information is available for S100PBPR. It was originally cloned from a pancreatic epithelioid carcinoma library and is located at 1p34.3. Despite a comprehensive search of the available data bases no sequence or structural homology to any other known protein could be detected. GFP-tagged S100PBPR was found to be located in the nucleus of HeLa cells where endogenous S100P is also present, suggesting that an interaction between these two proteins potentially occurs in the nucleus. S100P was also located in the cytoplasm, which was expected as it has recently been reported to be secreted.42 A cytoplasmic and nuclear distribution for GFP-S100P has previously been described in unstimulated A431 cells; stimulation with epidermal growth factor induced co-localization of GFP-S100P with an F-actin associated protein, ezrin, in microvillar membrane protrusions.43 Immunohistochemical analyses have also described nuclear and cytoplasmic immunoreactivity of S100P.32,39
S100PBPR is a widely expressed protein. By RT-PCR its expression could be detected in normal brain, breast, spleen, and lung. In addition in situ hybridization analysis showed the presence of S100PBPR transcript in pancreatic islets and at a lower level in a proportion of the acinar cell population. These findings are in agreement with expression information available for S100PBPR (FLJ12903, Hs.369253) on the Unigene website (http://www.ncbi.nlm.nih.gov/Unigene/), where S100PBPR expression is listed in a wide variety of normal and cancerous tissues, including purified pancreatic islets. Interestingly, in this study we also detected S100P by immunohistochemistry in the islets of normal pancreatic sections. These data suggest that in the healthy pancreas the expression patterns of S100P and S100PBPR are highly concordant, neither of the proteins being expressed in normal ductal cells.
Both S100P and S100PBPR were found by quantitative real-time PCR to be overexpressed in PanIN and PDAC samples compared to normal pancreata. Data for S100P were confirmed by immunohistochemistry, and were found to correlate significantly with increasing grade of PanIN lesions. Because an antibody for S100PBPR is not yet available, in situ hybridization was used to map the expression of S100PBPR in PDAC specimens. As for S100P, S100PBPR transcript was found to be located in the malignant PDAC cells. The similarity in the expression patterns of these two proteins, both in healthy pancreas and pancreatic cancer, provides support of a potential interaction between them, and their up-regulation in PanIN lesions indicates that they may both be involved in early pancreatic cancer development.
To date, there have been many studies describing the expression profiles of normal pancreas, CP, and PDAC samples;17–31,50 however, the analysis of PanINs has been hampered by the difficulty of obtaining sufficient material and number of these samples. Not all PanINs progress to malignant disease and it is hoped that analysis of these precursor lesions will result in the identification of genes that can be used to determine high-risk PanINs, ie, those likely to progress to invasive PDAC. Alterations in the sequence or expression levels of K-Ras, HER2/neu, p16INK4a, BRCA2, p53, DPC4, cyclin D1, and p21 have previously been described in these precursor lesions and a genetic progression model of PDAC is beginning to emerge.12,51,52
Although overexpression of S100P in PDAC has been well documented,20,23–26,31,32 in the current study we show S100P expression also in PanIN lesions. S100P was detected in only 2% of PanIN-1A but its expression increased significantly to 13% of PanIN-1B, 31% of PanIN-2, and 41% in PanIN-3 lesions. The percentage of positive cases was again found to increase significantly to 92% in invasive PDAC. These data allow us to add S100P to the existing model of pancreatic cancer progression. To date, K-Ras activation and overexpression of HER2/neu are the earliest changes in precursor lesions, being observed in PanIN-1A and PanIN-IB.7,53–56 These changes are closely followed by an increase in the number of p21-expressing lesions. Biankin and colleagues12 found 32% of PanIN-1B to be positive for p21; it is thought this may be a direct result of Ras activation or HER2/neu overexpression. Because S100P expression is observed in 13% of PanIN-1B it can be placed as one of the genes whose expression is altered very early in the development of PDAC. Loss of p16 is observed slightly later in the progression model, primarily in PanIN-2 and -3.8,9,57 Altered p53 expression, cyclin D1 elevation, decreased DPC4, and BRCA2 loss of heterozygosity are later aberrations and occur at a lower frequency.5,10–13,55,58 Expression of S100P was found to be twice as prevalent in invasive PDAC (92% of cases) as in PanIN-3 lesions (41% of cases), suggesting that S100P may play an important role in progression from PanINs to invasive PDAC. As a result, S100P could prove to be a valuable marker for the prediction of clinically relevant PanINs, ie, those lesions that are likely to progress and may therefore require intervention.
Acknowledgments
We thank Dr. Sam Lattimore for his assistance with the statistical analysis included in this study and Krishna Caulee for expert technical assistance.
Footnotes
Address reprint requests to Professor Nick R. Lemoine, Molecular Oncology Unit, Barts and The London School of Medicine and Dentistry, John Vane Science Centre, Charterhouse Square, London, EC1M 6BQ, UK. E-mail: nick.lemoine@cancer.org.uk.
Supported by Cancer Research UK, the National Translational Cancer Research Network, the Digestive Cancer Campaign, and the Mike Stone Cancer Research Fund.
S.E.D. and T.C.-J. contributed equally to the study.
References
- Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- Warshaw AL, Fernandez-del Castillo C. Pancreatic carcinoma. N Engl J Med. 1992;326:455–465. doi: 10.1056/NEJM199202133260706. [DOI] [PubMed] [Google Scholar]
- Neoptolemos JP, Stocken DD, Friess H, Bassi C, Dunn JA, Hickey H, Beger H, Fernandez-Cruz L, Dervenis C, Lacaine F, Falconi M, Pederzoli P, Pap A, Spooner D, Kerr DJ, Buchler MW. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004;350:1200–1210. doi: 10.1056/NEJMoa032295. [DOI] [PubMed] [Google Scholar]
- Kern S, Hruban R, Hollingsworth MA, Brand R, Adrian TE, Jaffee E, Tempero MA. A white paper: the product of a pancreas cancer think tank. Cancer Res. 2001;61:4923–4932. [PubMed] [Google Scholar]
- DiGiuseppe JA, Hruban RH, Goodman SN, Polak M, van den Berg FM, Allison DC, Cameron JL, Offerhaus GJ. Overexpression of p53 protein in adenocarcinoma of the pancreas. Am J Clin Pathol. 1994;101:684–688. doi: 10.1093/ajcp/101.6.684. [DOI] [PubMed] [Google Scholar]
- DiGiuseppe JA, Hruban RH, Offerhaus GJ, Clement MJ, van den Berg FM, Cameron JL, van Mansfeld AD. Detection of K-ras mutations in mucinous pancreatic duct hyperplasia from a patient with a family history of pancreatic carcinoma. Am J Pathol. 1994;144:889–895. [PMC free article] [PubMed] [Google Scholar]
- Day JD, Digiuseppe JA, Yeo C, Lai-Goldman M, Anderson SM, Goodman SN, Kern SE, Hruban RH. Immunohistochemical evaluation of HER-2/neu expression in pancreatic adenocarcinoma and pancreatic intraepithelial neoplasms. Hum Pathol. 1996;27:119–124. doi: 10.1016/s0046-8177(96)90364-0. [DOI] [PubMed] [Google Scholar]
- Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- Wilentz RE, Geradts J, Maynard R, Offerhaus GJ, Kang M, Goggins M, Yeo CJ, Kern SE, Hruban RH. Inactivation of the p16 (INK4A) tumor-suppressor gene in pancreatic duct lesions: loss of intranuclear expression. Cancer Res. 1998;58:4740–4744. [PubMed] [Google Scholar]
- Goggins M, Hruban RH, Kern SE. BRCA2 is inactivated late in the development of pancreatic intraepithelial neoplasia: evidence and implications. Am J Pathol. 2000;156:1767–1771. doi: 10.1016/S0002-9440(10)65047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, Kern SE, Hruban RH. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–2006. [PubMed] [Google Scholar]
- Biankin AV, Kench JG, Morey AL, Lee CS, Biankin SA, Head DR, Hugh TB, Henshall SM, Sutherland RL. Overexpression of p21(WAF1/CIP1) is an early event in the development of pancreatic intraepithelial neoplasia. Cancer Res. 2001;61:8830–8837. [PubMed] [Google Scholar]
- Luttges J, Galehdari H, Brocker V, Schwarte-Waldhoff I, Henne-Bruns D, Kloppel G, Schmiegel W, Hahn SA. Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. Am J Pathol. 2001;158:1677–1683. doi: 10.1016/S0002-9440(10)64123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Wallrapp C, Muller-Pillasch F, Micha A, Wenger C, Geng M, Solinas-Toldo S, Lichter P, Frohme M, Hoheisel JD, Adler G, Gress TM. Novel technology for detection of genomic and transcriptional alterations in pancreatic cancer. Ann Oncol. 1999;10:64–68. [PubMed] [Google Scholar]
- Crnogorac-Jurcevic T, Efthimiou E, Capelli P, Blaveri E, Baron A, Terris B, Jones M, Tyson K, Bassi C, Scarpa A, Lemoine NR. Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene. 2001;20:7437–7446. doi: 10.1038/sj.onc.1204935. [DOI] [PubMed] [Google Scholar]
- Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21:4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friess H, Ding J, Kleeff J, Fenkell L, Rosinski JA, Guweidhi A, Reidhaar-Olson JF, Korc M, Hammer J, Buchler MW. Microarray-based identification of differentially expressed growth- and metastasis-associated genes in pancreatic cancer. Cell Mol Life Sci. 2003;60:1180–1199. doi: 10.1007/s00018-003-3036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzmann R, Foerder M, Alldinger I, Staub E, Brummendorf T, Ropcke S, Li X, Kristiansen G, Jesnowski R, Sipos B, Lohr M, Luttges J, Ockert D, Kloppel G, Saeger HD, Pilarsky C. Gene expression profiles of microdissected pancreatic ductal adenocarcinoma. Virchows Arch. 2003;443:508–517. doi: 10.1007/s00428-003-0884-1. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–8622. [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- Sato N, Fukushima N, Matsubayashi H, Goggins M. Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene. 2004;23:1531–1538. doi: 10.1038/sj.onc.1207269. [DOI] [PubMed] [Google Scholar]
- Tan ZJ, Hu XG, Cao GS, Tang Y. Analysis of gene expression profile of pancreatic carcinoma using cDNA microarray. World J Gastroenterol. 2003;9:818–823. doi: 10.3748/wjg.v9.i4.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Hata F, Nishimori H, Honmou O, Yasoshima T, Nomura H, Ohno K, Hirai I, Kamiguchi K, Isomura H, Hirohashi Y, Denno R, Sato N, Hirata K. Differential gene expression screening between parental and highly metastatic pancreatic cancer variants using a DNA microarray. J Exp Clin Cancer Res. 2003;22:307–313. [PubMed] [Google Scholar]
- Yoshida K, Ueno S, Iwao T, Yamasaki S, Tsuchida A, Ohmine K, Ohki R, Choi YL, Koinuma K, Wada T, Ota J, Yamashita Y, Chayama K, Sato K, Mano H. Screening of genes specifically activated in the pancreatic juice ductal cells from the patients with pancreatic ductal carcinoma. Cancer Sci. 2003;94:263–270. doi: 10.1111/j.1349-7006.2003.tb01431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XJ, Long J, Fu DL, Zhang QH, Ni QX. Analysis of gene expression profiles in pancreatic carcinoma by using cDNA microarray. Hepatobiliary Pancreat Dis Int. 2003;2:467–470. [PubMed] [Google Scholar]
- Nakamura T, Furukawa Y, Nakagawa H, Tsunoda T, Ohigashi H, Murata K, Ishikawa O, Ohgaki K, Kashimura N, Miyamoto M, Hirano S, Kondo S, Katoh H, Nakamura Y, Katagiri T. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene. 2004;23:2385–2400. doi: 10.1038/sj.onc.1207392. [DOI] [PubMed] [Google Scholar]
- Crnogorac-Jurcevic T, Missiaglia E, Blaveri E, Gangeswaran R, Jones M, Terris B, Costello E, Neoptolemos JP, Lemoine NR. Molecular alterations in pancreatic carcinoma: expression profiling shows that dysregulated expression of S100 genes is highly prevalent. J Pathol. 2003;201:63–74. doi: 10.1002/path.1418. [DOI] [PubMed] [Google Scholar]
- Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Becker T, Gerke V, Kube E, Weber K. S100P, a novel Ca(2+)-binding protein from human placenta. cDNA cloning, recombinant protein expression and Ca2+ binding properties. Eur J Biochem. 1992;207:541–547. doi: 10.1111/j.1432-1033.1992.tb17080.x. [DOI] [PubMed] [Google Scholar]
- Sato N, Hitomi J. S100P expression in human esophageal epithelial cells: human esophageal epithelial cells sequentially produce different S100 proteins in the process of differentiation. Anat Rec. 2002;267:60–69. doi: 10.1002/ar.10085. [DOI] [PubMed] [Google Scholar]
- Guerreiro Da Silva ID, Hu YF, Russo IH, Ao X, Salicioni AM, Yang X, Russo J. S100P calcium-binding protein overexpression is associated with immortalization of human breast epithelial cells in vitro and early stages of breast cancer development in vivo. Int J Oncol. 2000;16:231–240. [PubMed] [Google Scholar]
- Bertram J, Palfner K, Hiddemann W, Kneba M. Elevated expression of S100P, CAPL and MAGE 3 in doxorubicin-resistant cell lines: comparison of mRNA differential display reverse transcription-polymerase chain reaction and subtractive suppressive hybridization for the analysis of differential gene expression. Anticancer Drugs. 1998;9:311–317. doi: 10.1097/00001813-199804000-00004. [DOI] [PubMed] [Google Scholar]
- Amler LC, Agus DB, LeDuc C, Sapinoso ML, Fox WD, Kern S, Lee D, Wang V, Leysens M, Higgins B, Martin J, Gerald W, Dracopoli N, Cordon-Cardo C, Scher HI, Hampton GM. Dysregulated expression of androgen-responsive and nonresponsive genes in the androgen-independent prostate cancer xenograft model CWR22–R1. Cancer Res. 2000;60:6134–6141. [PubMed] [Google Scholar]
- Mousses S, Bubendorf L, Wagner U, Hostetter G, Kononen J, Cornelison R, Goldberger N, Elkahloun AG, Willi N, Koivisto P, Ferhle W, Raffeld M, Sauter G, Kallioniemi OP. Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res. 2002;62:1256–1260. [PubMed] [Google Scholar]
- Terris B, Blaveri E, Crnogorac-Jurcevic T, Jones M, Missiaglia E, Ruszniewski P, Sauvanet A, Lemoine NR. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am J Pathol. 2002;160:1745–1754. doi: 10.1016/S0002-9440(10)61121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JM, Iannettoni MD, Orringer MB, Hanash S. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via RAGE. J Biol Chem. 2003;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- Koltzscher M, Neumann C, Konig S, Gerke V. Ca2+-dependent binding and activation of dormant ezrin by dimeric S100P. Mol Biol Cell. 2003;14:2372–2384. doi: 10.1091/mbc.E02-09-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek A, Jastrzebska B, Nowotny M, Kuznicki J. CacyBP/SIP, a calcyclin and Siah-1-interacting protein, binds EF-hand proteins of the S100 family. J Biol Chem. 2002;277:28848–28852. doi: 10.1074/jbc.M203602200. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- Kloppel G, Luttges J. WHO-classification 2000: exocrine pancreatic tumors. Verh Dtsch Ges Pathol. 2001;85:219–228. [PubMed] [Google Scholar]
- Koltzscher M, Gerke V. Identification of hydrophobic amino acid residues involved in the formation of S100P homodimers in vivo. Biochemistry. 2000;39:9533–9539. doi: 10.1021/bi000257+. [DOI] [PubMed] [Google Scholar]
- Gribenko AV, Makhatadze GI. Oligomerization and divalent ion binding properties of the S100P protein: a Ca2+/Mg2+-switch model. J Mol Biol. 1998;283:679–694. doi: 10.1006/jmbi.1998.2116. [DOI] [PubMed] [Google Scholar]
- Gribenko AV, Guzman-Casado M, Lopez MM, Makhatadze GI. Conformational and thermodynamic properties of peptide binding to the human S100P protein. Protein Sci. 2002;11:1367–1375. doi: 10.1110/ps.0202202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- Hruban RH, Wilentz RE, Kern SE. Genetic progression in the pancreatic ducts. Am J Pathol. 2000;156:1821–1825. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern SE, Hruban RH, Hidalgo M, Yeo CJ. An introduction to pancreatic adenocarcinoma genetics, pathology and therapy. Cancer Biol Ther. 2002;1:607–613. doi: 10.4161/cbt.307. [DOI] [PubMed] [Google Scholar]
- Caldas C, Hahn SA, Hruban RH, Redston MS, Yeo CJ, Kern SE. Detection of K-ras mutations in the stool of patients with pancreatic adenocarcinoma and pancreatic ductal hyperplasia. Cancer Res. 1994;54:3568–3573. [PubMed] [Google Scholar]
- Terhune PG, Phifer DM, Tosteson TD, Longnecker DS. K-ras mutation in focal proliferative lesions of human pancreas. Cancer Epidemiol Biomarkers Prev. 1998;7:515–521. [PubMed] [Google Scholar]
- Apple SK, Hecht JR, Lewin DN, Jahromi SA, Grody WW, Nieberg RK. Immunohistochemical evaluation of K-ras, p53, and HER-2/neu expression in hyperplastic, dysplastic, and carcinomatous lesions of the pancreas: evidence for multistep carcinogenesis. Hum Pathol. 1999;30:123–129. doi: 10.1016/s0046-8177(99)90265-4. [DOI] [PubMed] [Google Scholar]
- Luttges J, Schlehe B, Menke MA, Vogel I, Henne-Bruns D, Kloppel G. The K-ras mutation pattern in pancreatic ductal adenocarcinoma usually is identical to that in associated normal, hyperplastic, and metaplastic ductal epithelium. Cancer. 1999;85:1703–1710. [PubMed] [Google Scholar]
- Hansel DE, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Annu Rev Genom Hum Genet. 2003;4:237–256. doi: 10.1146/annurev.genom.4.070802.110341. [DOI] [PubMed] [Google Scholar]
- Boschman CR, Stryker S, Reddy JK, Rao MS. Expression of p53 protein in precursor lesions and adenocarcinoma of human pancreas. Am J Pathol. 1994;145:1291–1295. [PMC free article] [PubMed] [Google Scholar]