Figure 1.
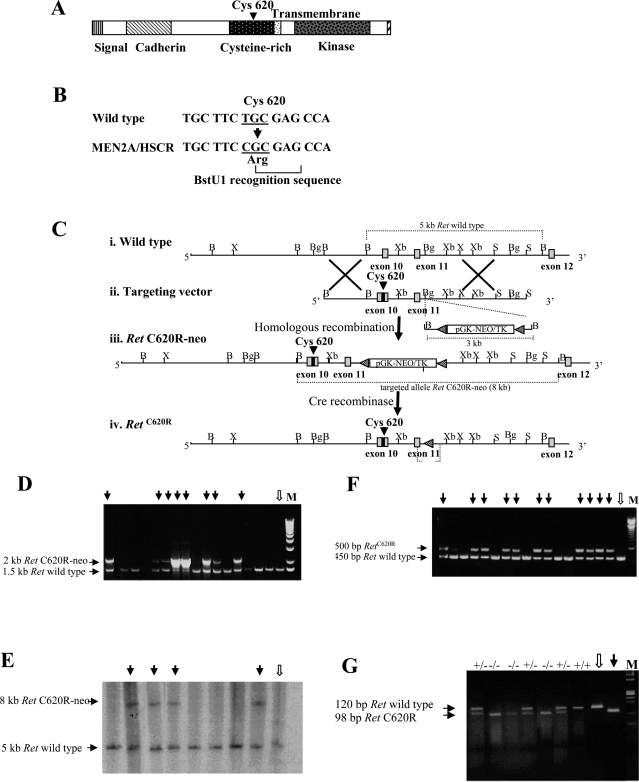
Introducing a MEN 2A/HSCR-associated mutation into the Ret locus. A: Structure of the normal protein encoded by the Ret gene. The location of the Cys620 is indicated. B: Site-directed mutagenesis to introduce the Cys620Arg mutation that generates a BstU1 recognition sequence (CGCG). C: Schematic representation of the targeting strategy. i: A fragment of the normal Ret gene including exons 10 to 12 (gray boxes). ii: The targeting vector includes a segment of genomic DNA containing exons 10 and 11. Exon 10 contains the mutation at position 620 so that the Cys in the Ret protein is replaced by an Arg. A selection cassette containing the neo gene flanked by two lox P sites has been inserted into the BglII site in the intron adjacent to exon 11. Lox P sites are indicated by black triangles. iii: Homologous recombination between the Ret genomic locus and the targeting construct introduced the mutant exon 10 and the loxP-neo-loxP gene in the intron adjacent to exon 11, to produce the RETC620R-neo allele. iv: The RetC620R allele was produced by Cre-mediated excision of the selection cassette. The restriction enzyme sites shown are B, BamHI; Bg, BglII; S, SacI; X, XhoI; and Xb, XbaI. D: PCR-based screening for the analysis of targeted ES cell clones. DNA extracted from G418-resistant ES cell clones was amplified by PCR (P1, 5′-aagctctctagtcgaggaat-3′ and P2, 5′-ggcctctgaaggactgaa-3′) to produce both a 2-kb fragment corresponding to the insertion in one allele of targeted clones (black arrows) and/or a 1.5-kb fragment in the wild-type ES cells (white arrow) and in the nontargeted clones. E: PCR results were confirmed by Southern blot analysis in which BamHI-digested DNA was hybridized to a probe containing exon 11 and therefore present in both targeted and nontargeted clones. The probe was obtained by amplifying across the region using PCR (P3, 5′-acagggggaggtggtacagt-3′ and P4, 5′-atgccgtatccaccatctgt-3′). In this confirmatory screen the expected size of the targeted allele was 8 kb (black arrows), compared to 5 kb for the wild-type allele (white arrow). The position of the two alleles is indicated in C, iii and i, respectively. F: PCR-based analysis of targeted clones after Cre-mediated recombination. Targeted clones that had undergone Cre-mediated excision of the selection cassette displayed a band of the expected size (black arrows) after DNA amplification across the region of the inserted cassette. As a loxP site remains in the targeted allele this PCR amplification across the region (P5, 5′-cagggcttcccaatcagtt-3′ and P6, 5′-ccgagtacatgtgtgcctgt-3′) yields a band of ∼500 bp, which is ∼50 bp larger than the wild-type (white arrow) or nontargeted alleles. The location of the P5 and P6 primers is indicated in C, iv. G: PCR-based genotyping of tail biopsies from a representative litter of pups from a heterozygous breeding pair. A 120-bp amplification product (P7, 5′-tgccgacattgttggaggaac-3′ and P8, 5′-cctggctgttcctctggct-3′) was generated from the wild-type allele, which is not cleaved after digestion with the restriction enzyme BstUI. Conversely, amplification and BstUI digestion produced a product corresponding to 98 bp for the allele bearing the targeted C620R substitution. The products of the amplification and restriction enzyme digestion of wild-type DNA (white arrow) and DNA from the targeted RetC620R vector (black arrow) were loaded on the gel as internal controls. The genotype of each pup is indicated. +/+ wild-type; +/− heterozygous; −/− homozygous Ret620 mutant.