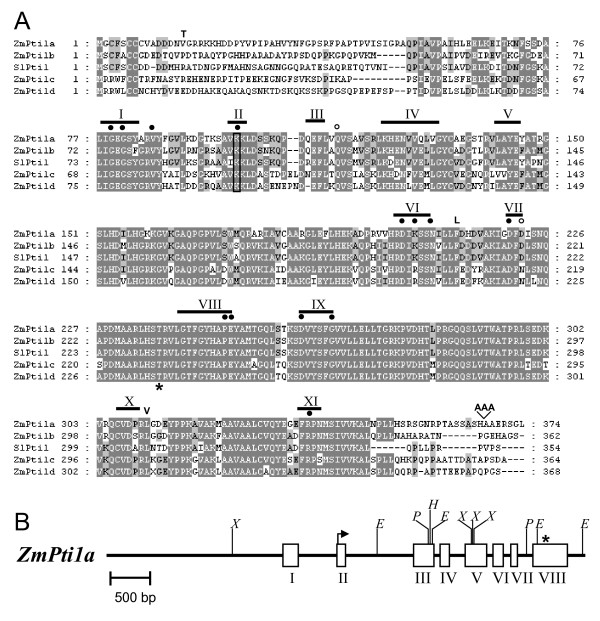
Figure 1.
Similarity and predicted genomic structure of ZmPti1a. (A) Alignment of Pti1 kinases from maize with SlPti1 from tomato. Amino acids identical in at least three of the sequences are highlighted in grey. The 11 canonical subdomains conserved in serine/threonine kinases are indicated with Roman numerals. Invariant residues common to the majority of protein kinases are marked with black dots. Invariant residues that are conserved in other protein kinases but not in Pti1 kinases are marked with open circles. The highly conserved lysine residue in subdomain II which is required for activity in SlPti1 and most protein kinases is boxed. Threonine 233 has been identified as the major site of SlPti1 phosphorylation by SlPto and is marked with an asterisk. Amino acids which differ between ZmPti1a and the deduced protein sequence of the second cloned ZmPti1a cDNA [GenBank:AY554282] are indicated above the sequences. (B) Genomic locus and restriction map of the ZmPti1a gene. Exons are indicated as boxes with Roman numerals. Start and stop of the open reading frame are marked with an arrow and asterisk, respectively. E, EcoRI; H, HindIII; P, PstI, X, XhoI.