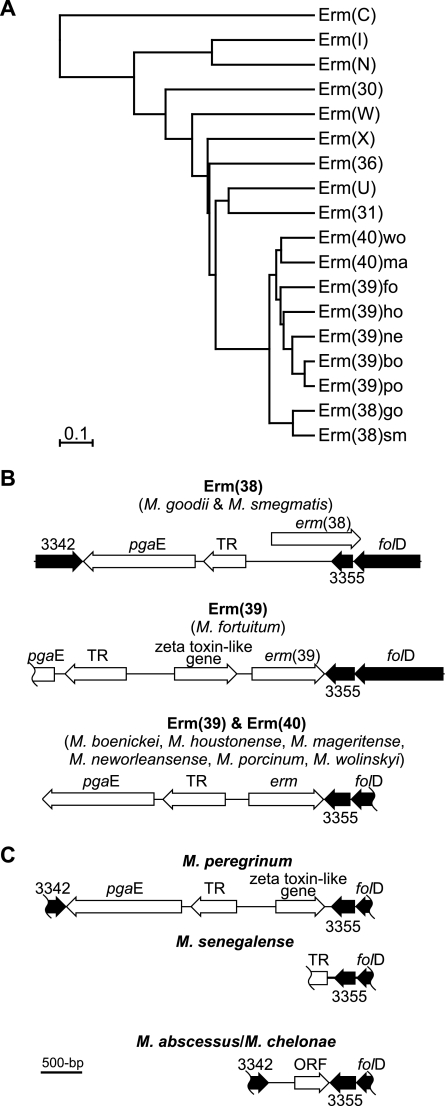
FIG. 1.
(A) Phylogram showing the predicted relationships of the RGM Erm proteins, with the representative alleles of the eight most closely related Erm classes, and rooted on Erm(C) of Staphylococcus aureus (GenBank accession no. AAA20192). The Erm class representatives (with GenBank accession numbers) are Erm(30) of Streptomyces venezuelae (AAC69328), Erm(31) of S. venezuelae (AAC69327), Erm(36) of Micrococcus luteus (AAL68827), Erm(I) of Streptomyces mycarofaciens (A60725), Erm(N) of Streptomyces fradiae (CAA66307), Erm(U) of Streptomyces lincolnensis (CAA55770), Erm(W) of Micromonospora griseorubida (P43433), and Erm(X) of Corynebacterium jeikeium (AAK28910). For the RGM Erm proteins, Erm(38), Erm(39), and Erm(40), the allele (or RGM species) is indicated by the letters after the right parenthesis: bo, M. boenickei; fo, M. fortuitum; go, M. goodii; ho, M. houstonense; ma, M. mageritense; ne, M. neworleansense; po, M. porcinum; sm, M. smegmatis; and wo, M. wolinskyi. The scale bar shows the evolutionary distance (i.e., the amount of divergence, or time since splitting, from a theoretical common ancestor). (B and C) The genetic organization of the folD regions of erm gene-positive and erm gene-negative RGM, respectively. The shaded genes are similar (>64% amino acid identity) to M. tuberculosis strain H37Rv genes Rv3342 (GenBank accession no. CAA17114), Rv3355c (GenBank accession no. NP_217872), and folD (GenBank accession no. NP_217873). Other genes are those encoding TR, a putative tetR family transcriptional regulator, and pgaE, a putative polyketide synthase, and zeta toxin-like, containing a zeta toxin conserved domain (Pfam06414) and ∼50% amino acid identity to conserved hypothetical genes of Polaromonas spp. (GenBank accession no. EAM36411) and Xanthomonas oryzae (GenBank accession no. AAW75612). ORF, open reading frame with a likely ribosome binding site (AAGGGA).