Abstract
CelB (BH0603) from Bacillus halodurans is a modular glycoside hydrolase with a family 5 catalytic module, an immunoglobulin-like module, and module PfamB of unknown function. The recombinant PfamB module bound to Avicel and was essential for CelB hydrolytic function. We propose that module PfamB be designated a new carbohydrate-binding module.
Some cellulases have a modular architecture composed of catalytic modules appended to auxiliary modules; for example, carbohydrate-binding modules (CBMs) provide the targeting function that delivers catalytic modules to substrates (2, 3). Forty-five CBM families are defined in the CAZy database (http://afmb.cnrs-mrs.fr/CAZY/). Encoded at locus BH0603 (GenBank accession no. BA000004) in the genome of the alkaliphilic bacterium Bacillus halodurans (7) is a putative modular endo-β-1,4-glucanase (CelB) composed of a glycoside hydrolase family 5 (GHF5) catalytic module, an immunoglobulin (Ig)-like module, and module PfamB of unknown function (Fig. 1). We aimed to ascertain the function of module PfamB, whose alignment (8) with select PfamB-type modules is presented in Fig. 2.
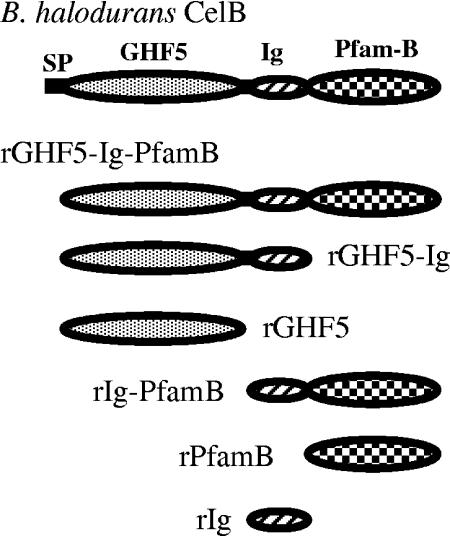
FIG. 1.
Modular organization of CelB and its truncated derivatives. CelB consists of a signal peptide (SP), a GHF5 module, an Ig-like module, and a PfamB module.
FIG. 2.
Alignment of amino acid sequences of Pfam-B modules from B. halodurans CelB (Bh), B. agaradhaerens cellulase (Ba), P. lautus putative CelB (Pl), and a cellulase of an uncultured bacterium (UB). The conserved amino acid residues are indicated by an asterisk below the alignment. The numbers at the start and end of the respective sequence lines refer to amino acid residues in the parent proteins (B. halodurans [accession no. BA000004], B. lautus [accession no. A28172], B. agaradhaerens [accession no. AJ537597], and an uncultured bacterium [accession no. AJ537596]).
A PCR-based deletion method was used to construct six-histidine-tagged recombinant CelB derivatives (Fig. 1). B. halodurans genomic DNA was used as the PCR template (5). The primers used for PCR amplification of the portions of celB encoding CelB amino acids 26 to 574 (rGHF5-Ig-PfamB), 26 to 456 (rGHF5-Ig), 26 to 345 (rGHF5), 346 to 574 (rIg-PfamB), 346 to 456 (rIg), and 457 to 574 (rPfamB) were as follows: forward, CelBF (5′-GGATCCGTTAGTTCTGCTCATGAGGATGTG-3′; rGHF5-Ig-PfamB), CmF (5′-CCGCCATGGGCGCTCATGAGGATGTGA-3′; rGHF5-Ig and rGHF5), DufXF (5′-TTGGATCCTGGCATACGTACGAATGG-3′; rIg-PfamB), XF (5′-TGGGATCCTATCGTACGCCTGTATTGC-3′; rPfamB), and DufF (5′-AGCATTTCAATCCCATGGGCTACGAATGGT-3′; rIg); reverse, CelBR (5′-GTCGACATTCGGGTAACACCATAGAAAGC-3′; rGHF5-Ig-PfamB), DufR (5′-ATACAGGCGTCTCGAGCGTATTCACCCGAA-3′; rGHF5-Ig and rIg), CmR (5′-TCATACCACTCGAGCGTATGACGAT-3′; rGHF5), DufXR (5′-GTCGACTTCGGGTAACACCATAGAAAGC-3′; rIg-PfamB), and XR (5′-GTCGACGGGTAACACCATAGAAAGCGCTT-3′; rPfamB). Primers incorporated BamHI or NcoI restriction sites (underlined boldface nucleotides) at the 5′ end and SalI or XhoI restriction sites at the 3′ end. Thermal cycling conditions were 1 cycle of initial denaturation at 98°C for 5 min; 26 cycles of denaturation, annealing, and extension at 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min, respectively; and 1 final extension cycle of 7 min at 72°C.
PCR amplicons were inserted into the pET-28a vector (Novagen) to generate desired vector-insert cassettes for recombinant-protein production after transformation into Escherichia coli BL21(DE3) (6). Luria-Bertani (LB) broth was inoculated with E. coli transformants and incubated in shake flasks to an optical density at 600 nm of ∼0.6. Isopropyl-β-d-thiogalactopyranoside was added to a concentration of 1 mM, and the culture was incubated for a further 12 h at 20°C. Cells were collected by centrifugation (4,200 × g, 10 min), and the pellet was resuspended in 7 ml lysis buffer (50 mM NaH2PO4, 0.3 M NaCl, 10 mM imidazole, pH 8.0). Cells were disrupted by sonication and centrifuged, and the supernatant was collected. Recombinant polypeptides were purified from soluble protein extracts with Ni-nitrilotriacetic acid spin columns (QIAGEN) according to the manufacturer's instructions. All purified proteins showed single bands in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (4 and data not shown). Protein concentration was determined with the Bradford reagent with bovine serum albumin (BSA) as the standard.
Binding of CelB derivatives to Avicel was determined qualitatively and visualized by Coomassie blue staining following SDS-PAGE. Purified protein (30 μg) was mixed with 10 mg of Avicel in a final volume of 200 μl of 5 mM Tris-HCl buffer, pH 8.9. Tubes were incubated on ice at 4°C for 1 h with regular gentle mixing before being centrifuged (12,000 × g, 2 min), and the supernatant, containing unbound protein, was carefully removed. The cellulose pellet was then washed in 200 μl of phosphate-buffered saline before being resuspended in 50 μl of SDS-PAGE buffer and boiled for 10 min to dissociate any bound protein. Controls with (i) protein but no Avicel and (ii) BSA with Avicel were included to ensure (i) that no precipitation occurred during the assay period and (ii) the efficiency of the washing step. Bound and unbound protein fractions were analyzed by 12.5% (rGHF5-Ig-PfamB) and 18% (rIg-PfamB, rIg, and rPfamB) SDS-PAGE (4). Analysis of protein binding to soluble substrates was by affinity electrophoresis (9). Native-polyacrylamide gels, without and with binding substrate at a concentration of 0.25% (wt/vol) added prior to polymerization, were prepared separately and run in parallel at 4°C. BSA was used as a noninteracting negative control.
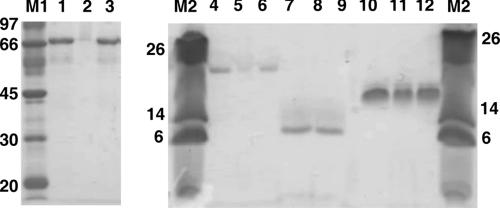
Figure 3 shows the profile of CelB derivative binding to Avicel. rGHF5-Ig-PfamB and rIg-PfamB were detected only in the bound fraction, unlike rPfamB, which was detected in both the wash and bound fractions. Since an equal amount of protein was used for binding assays and an equal volume of sample was loaded per gel well, it is unlikely that rPfamB in the wash fraction was due to release of loosely bound protein from overloaded Avicel. rPfamB may have bound the Avicel loosely and a part of the bound protein was dislodged from the Avicel by washing. Thus, rIg-PfamB bound relatively tightly to Avicel compared to rPfamB. Considering that rIg failed to bind Avicel, the above observation may be attributed to the separation of the Ig-like module from rIg-PfamB. Binding of rGHF5-Ig-PfamB to soluble substrates such as carboxymethyl cellulose (CMC) and barley β-glucan (affinity electrophoresis data not shown) was too weak to reliably make inferences.
FIG. 3.
Binding of CelB derivatives to Avicel as assessed by SDS-PAGE. The sizes (in kilodaltons) of molecular mass standards are indicated to the left of each gel. Lanes: M1, molecular mass marker; 1, untreated r 4, untreated rIg-PfamB; 5; unbound rIg-PfamB; 6, bound rIg-PfamB; 7, untreated rIg; 8, unbound rIg; 9, bound rIg; 10, untreated rIg-PfamB; 11, unbound rIg-PfamB; 12, bound rIg-PfamB.
Hydrolytic activity of CelB derivatives was determined as follows. Recombinant protein samples (30 μg protein) were incubated with 1% (wt/vol) Avicel (Merck), CMC (low viscosity; Sigma), hydroxyethyl cellulose (Fluka), barley β-glucan (Sigma), birchwood xylan (Sigma), and laminarin (Nacalai Tesque, Kyoto, Japan) as substrates in a total volume of 1 ml of 50 mM Britton-Robinson's buffer (50 mM phosphoric acid, 50 mM boric acid, 50 mM acetic acid, pH 8.9). Liberated reducing sugars were assayed by the Somogyi-Nelson method (10). Specific activities of CelB derivatives on a range of substrates are shown in Table 1. It is generally accepted that a catalytic module and a CBM in the same polypeptide can function independently. Thus, artificial separation of the CBM from the catalytic module is not usually expected to affect the enzyme activity of the catalytic module toward soluble substrates (1). However, deletion of CelB C-terminal modules PfamB and Ig, both of which were devoid of any catalytic activity, led to significant changes in hydrolytic activity against all of the test substrates (Table 1). This phenomenon could not be attributed solely to the lack of physical binding because rGHF5-Ig-PfamB showed no binding affinity for soluble test substrates at all. It is probable that, along with the removal of module PfamB, interdomain interactions contributing crucially to the active-site structure of CelB were lost or disrupted. Module PfamB, which binds to Avicel, forms a significant part of the CelB polypeptide, without which the core enzyme has very limited overall action on cellulosic substrates. We propose that module PfamB be classified as a CBM.
TABLE 1.
Activities of rGHF5-Ig-PfamB, rGHF5-Ig, and rGHF5 toward various polysaccharides
| Substrate | Sp act (relative activity)a
|
||
|---|---|---|---|
| rGHF5-Ig-PfamB | rGHF5-Ig | rGHF5 | |
| β-Glucan | 1,294.5 (100) | 52.3 (4.0) | 5.0 (0.2) |
| Hydroxyethyl cellulose | 515.1 (39.1) | 10.1 (0.8) | 1.5 (0.1) |
| CMC | 130.9 (10.1) | 19.6 (1.5) | 2.0 (0.2) |
| Avicel | 0.012 (0.001) | NDb (—c) | ND (—) |
| Laminarin | ND (—) | ND (—) | ND (—) |
| Birchwood xylan | ND (—) | ND (—) | ND (—) |
Specific-activity assays were performed at 40°C in Britton-Robinson's buffer (pH 8.9), and the values shown are micromoles of glucose or xylose generated per minute per milligram of protein. Relative activities are expressed as percentages of the specific activity of GHF5-Ig-PfamB toward barley β-glucan.
ND, no activity detected.
—, not determined.
Acknowledgments
We gratefully acknowledge the financial support given by the Research Institute of Innovative Technology for the Earth (RITE) of the Ministry of Economy, Trade, and Industry (METI) of Japan and by the National Research Council of Thailand under the Thai-Japan (Japan Science and Technology Agency) Cooperative Research Program.
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.Arai, T., R. Araki, A. Tanaka, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2003. Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to cellulose hydrolysis. J. Bacteriol. 185:504-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davie. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hildén, L., and G. Johansson. 2004. Recent developments on cellulases and carbohydrate-binding modules with cellulose affinity. Biotechnol. Lett. 26:1683-1693. [DOI] [PubMed] [Google Scholar]
- 4.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 5.Saito, H., and K. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 6.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, P. R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomme, P., A. Boraston, J. M. Kormos, R. A. J. Warren, and D. G. Kilburn. 2000. Affinity electrophoresis for the identification and characterization of soluble sugar binding by carbohydrate-binding modules. Enzyme Microb. Technol. 27:453-458. [DOI] [PubMed] [Google Scholar]
- 10.Wood, T. M., and K. M. Bhat. 1988. Methods in measuring cellulase activities. Methods Enzymol. 160:87-112. [Google Scholar]