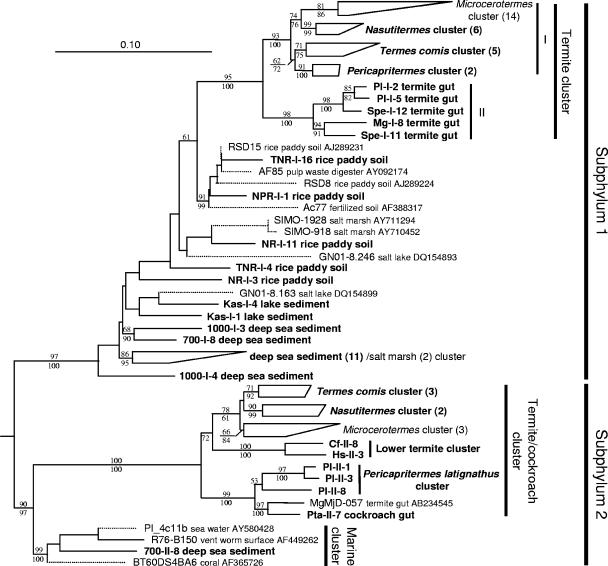
FIG. 5.
Phylogenetic tree showing the relationship of the 16S rRNA phylotypes affiliated with the candidate phylum TG3. An ML tree was constructed as the framework using the fastDNAml program implemented in ARB. The tree topology was basically congruent with the ML trees constructed using the PHYML program with the GTR or GTR+G+I model. An ME tree was also constructed with the GTR+G+I model. A total of 1,007 unambiguously aligned nucleotides were used, corresponding to positions 165 to 1225 in E. coli (J01695). Some phylotypes of the subphylum 1 were obtained by PCR using the primer set designed specific to the subphylum 2. This was caused by unexpected matching of the forward primer for subphylum 2 to some of the subphylum 1 phylotypes that had not been obtained before this study. Bootstrap tests were performed with 100 resamplings for both the ML tree with the GTR model and the ME tree, and the confidence values are indicated above (ML) and below (ME) the branches. The phylotypes and clusters obtained in this study are shown in bold letters. The short sequences (connected by dotted lines) found in public databases were added later by means of the ARB parsimony tool without changing the overall topology. The number of contained phylotypes in the compressed clusters are shown in parentheses. The origin of phylotypes are indicated in the clone codes as abbreviations listed in Table 2 for termites and Table 3 for other environments. Chlorobium limicola (Y10640) and Prosthecochloris vibrioforme (Y10649) were used as the outgroups. The full tree before the short sequences were added is published as Fig. S2 in the supplemental material. Pl, P. latignathus; Spe, Speculitermes sp.; Mg, M. gilvus; Cf, C. formosanus; Hs, H. sjoestedti; TNR, Tainan, Taiwan; NPR, Niigata, Japan; NR, Nagano, Japaan; Kas, Kasumi-ga-ura, Japan; 700, 700 m depth, Toyama, Japan; 1,000, 1,000 m depth, Toyama, Japan.