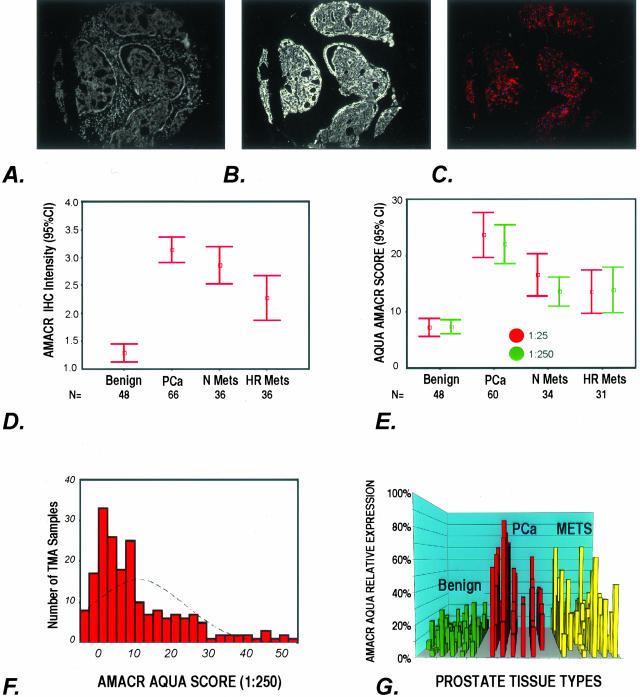
Figure 3.
A–G: AMACR expression as determined by AQUA and standard pathology review. A to C demonstrate how AQUA identifies prostate cancer from benign tissue in a tissue microarray sample (0.6 mm diameter). A: Using DAPI staining, all nucleated cells in both the stroma and epithelial compartments are identified. The cytokeratin-positive compartment identifies all epithelial cells (B) and Cy-5 expression (red) demonstrates the area of AMACR expression (C). The staining area and intensity of each compartment can be calculated. The AMACR-positive area, which overlaps with the cytokeratin mask is equivalent to the prostate cancer compartment. D: AMACR (p504s) protein expression by standard pathology review using immunohistochemistry. The expression of AMACR (p504s) was determined using the prostate cancer progression tissue microarray. Expression was scored as negative (score = 1), weak (score = 2), moderate (score = 3), and strong (score = 4). The results for each tissue type are presented as error bars of AMACR (p504s) protein expression with 95% confidence intervals. E: AQUA evaluation of AMACR expression using the prostate cancer progression tissue microarray at two antibody concentrations. AQUA measured similar AMACR protein expression levels for the different tissue categories [ie, benign, localized prostate cancer (PCa), metastatic prostate cancer to regional lymph nodes (Mets), and distant hormone-refractory metastatic prostate cancer (HR Mets)] at standard antibody concentrations (1:25) and at a concentration that would not be visible using standard immunohistochemistry (1:250). The intensity is corrected for area for each tissue microarray spot and measured using arbitrary units. The errors bars demonstrate that within 95% confidence intervals, benign prostate tissue does not score greater than 10 units. F: AMACR protein expression histogram for AQUA analysis. Using the arbitrary unit, this histogram demonstrates a relatively normal distribution of AMACR protein expression using samples from the prostate progression chip. This demonstrates the ability of the AQUA system to give a continuous readout of protein expression. G: Variable AMACR protein expression is appreciated by presenting the individual staining intensities for each sample based on tissue category. Although AMACR protein expression for the clinically localized prostate tumors is significantly higher than the population of benign samples, AMACR expression is still observed in benign tissues and can also be found to be low in other categories.