Figure 1.
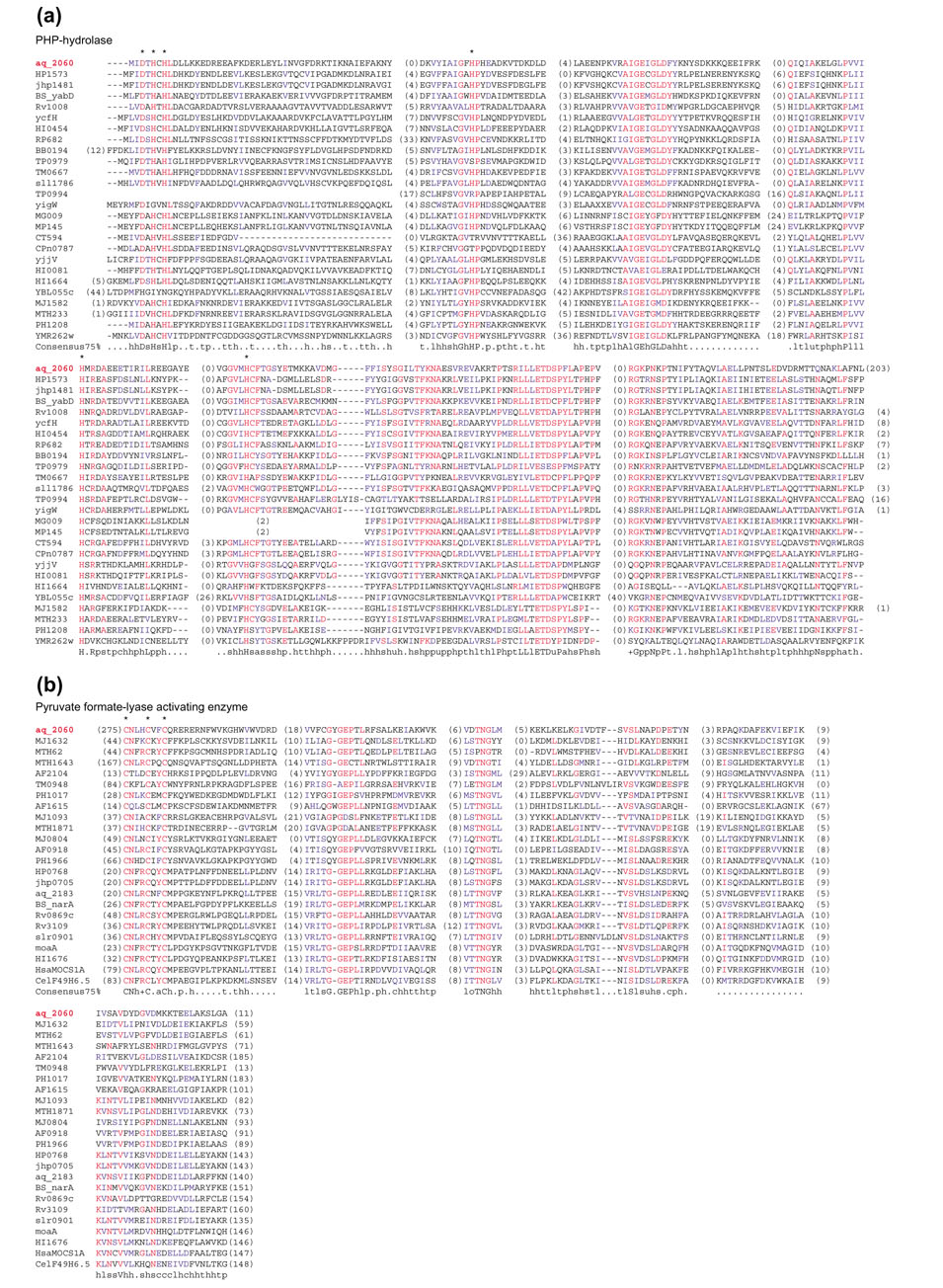
Multiple alignments of two domains comprising an interkingdom domian fusion. Alignments of (a) the PHP-hydrolase domain [4] and (b) the pyruvate formate lyase activating enzyme domain of the IKF protein aq_2060 from A. aeolicus. The sequences of the aq_2060 domains are placed with the most similar sequences of the corresponding stand-alone enzymes, bacterial ones in the case of PHP-hydrolase and archaeal ones in the case of the pyruvate formate lyase activating enzyme. The phylogenetic trees produced form these alignments are shown in Figure 2c. The numbers in parentheses show the lengths of regions between the aligned blocks that are not shown. The consensus includes amino acid residues and residue classes that are conserved in 75% of the aligned sequences; the residue classes are as follows: h, hydrophobic; l, aliphatic; a, aromatic; s, small; u, tiny; p, polar; b, big; t, residues with high turn-forming propensity. Asterisks show the predicted active site residues; note the replacements in some of the sequences that are predicted to be inactivated versions of the respective enzymes (see text). The alignments were colored using the BOXSHADE program [30]; individual residues conserved in at least 50% of the aligned sequences are in red; residues similar to the conserved ones and groups of conserved similar residues are in blue.
