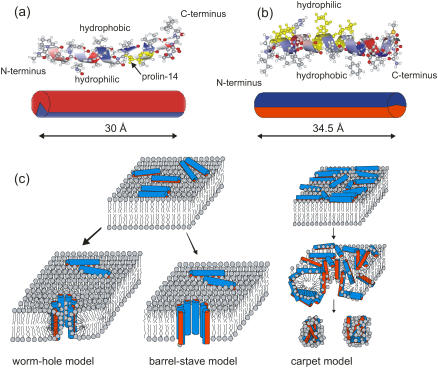
FIGURE 1.
(a) The α-helical conformation of alamethicin with its hydrophobic and hydrophilic sides of the helix (one of three crystal structures of alamethicin, the coordinate file 1AMT.PDB is taken from the Protein DataBank: http://www.rcsb.org/pdb), along with the representation as a amphiphilic cylinder, according to Fox and Richards (16) (bottom); the length of the molecule in this conformation is ∼30 Å. (b) The α-helical conformation of magainin with its hydrophilic and hydrophobic sides of the helix (one of 10 helical structures of magainin-2, the coordinate file 2MAG.PDB is taken from the Protein DataBank: http://www.rcsb.org/pdb), along with the representation as an amphiphilic cylinder, according to Gesell et al. (73) (bottom); the length of the molecule in this conformation is ∼34.5 Å. (c) Sketch of different models describing the functional mechanisms and underlying structure of antimicrobial peptides interacting with lipid bilayers, as discussed in the literature (74,2,4,75). (Left) The worm-hole model as proposed for magainin. (Top) Surface (S) state of antimicrobial peptides with the hydrophobic side groups anchored in the hydrophobic core of the bilayer. (Center) Barrel-stave model, as proposed for alamethicin. (Right) Carpet model: antimicrobial peptides crowding in the S state, and leading subsequently to micellation.