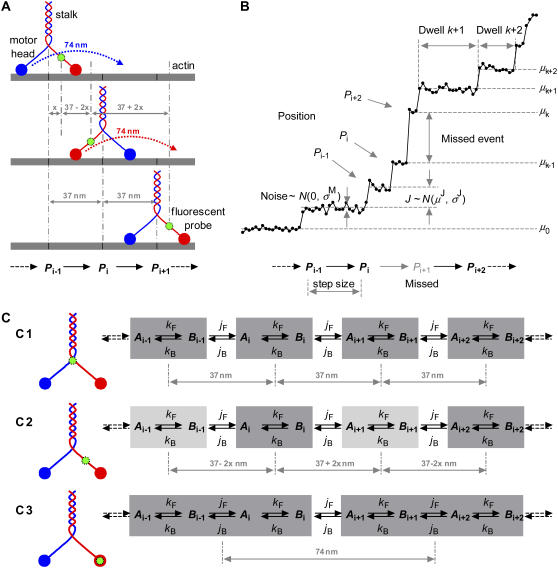
FIGURE 1.
Modeling the mechanochemistry of a molecular motor. (A) Myosin walks hand-over-hand along the actin filament, with the stalk taking 37 nm steps per ATP. The motor alternately swings its heads to walk: the rear head moves 74 nm (twice the stalk movement) and becomes the leading head. (B) Staircase data from single molecule measurements. Each data point is the position of the fluorescent probe measured along the axis of the filament, as a function of time. The motor may take more than one step within the sampling interval (notice the missed event between positions Pi and Pi+2). (C) The position of the fluorescent probe within the motor protein results in different step patterns. The mechanochemistry is modeled as an infinite chain of identical reaction units. At least two kinetic states per unit are necessary to describe the ATP binding step and the position translocation.