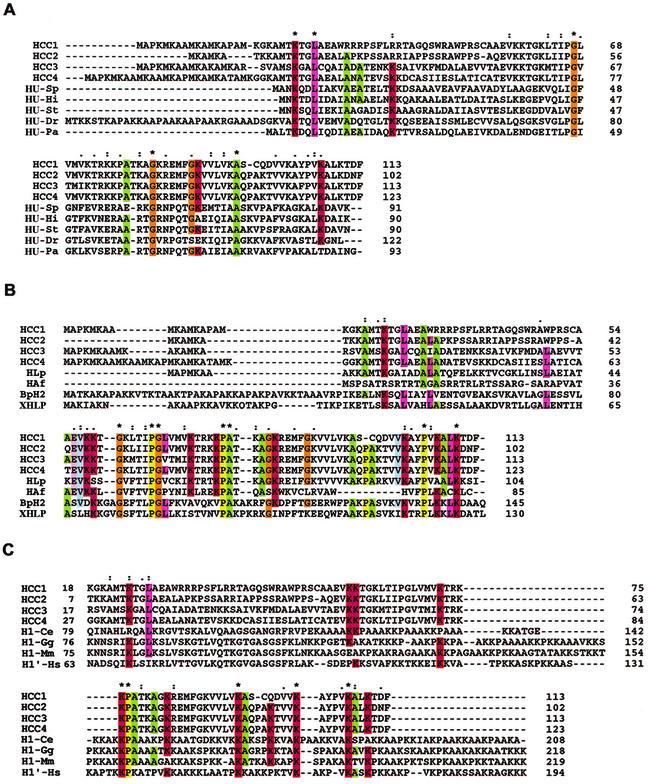
FIG. 1.
Alignment of HCCs with HUs, histone-like proteins, and histone H1. (A) Multiple alignments of HCCs and selective bacterial HUs: HCC1 (A56581), HCC2 (B56581), HCC3 (AY128510), and HCC4 (AY128511); HU-Pa (Pseudomonas aeruginosa, JC4061); HU-Dr (Deinococcus radiodurans, Q9RZ89); HU-St (Salmonella enterica serovar Typhimurium, S01732); HU-Hi (Haemophilus influenzae, P43722); and HU-Sp (Streptococcus pneumoniae, AAK75224). (B) Multiple alignments of dinoflagellate and bacterial histone-like proteins: HLP (L. polyedrum, AF482694), HAF (A. fundyense, AAD20317), BpH2 (B. pertussis, AAB40156), and XHLP (X. fastidiosa 9a5c, AAF84745). (C) Multiple alignments of HCCs and selective eukaryotic histone H1 proteins: H1-Ce (Caenorhabditis elegans, NP_510410), H1-Gg (Gallus gallus, P09987), H1′-Hs (Homo sapiens, XP_009973), and H1-Mm (Mus musculus, AAA37814). Asterisks indicate positions which have a single, fully conserved residue; colons indicate that one of the “strong” groups in parentheses is fully conserved (STA/NEQK/NHQK/NDEQ/QHRK/MILV/MILF/HY/FYW); periods indicate that one of the “weaker” groups in parentheses is fully conserved (CSA/ATV/SAG/STNK/STPA/SGND/SNDEQK/NDEQHK/NEQHRK/FVLIM/HFY).