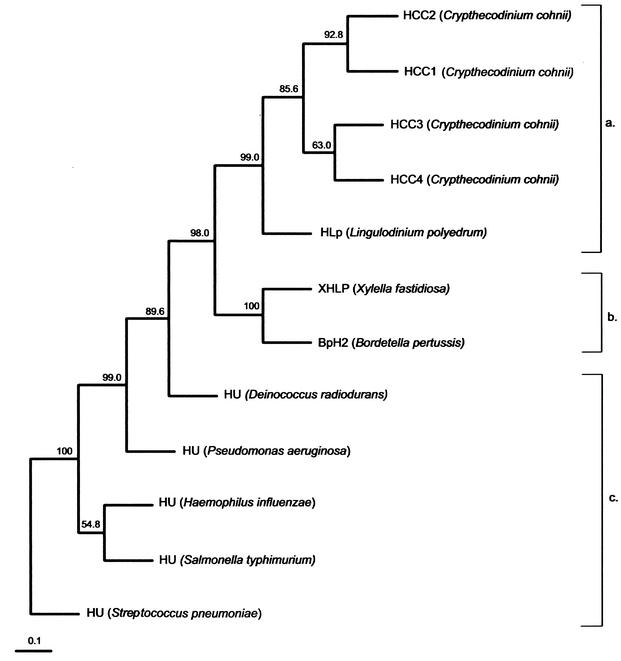
FIG. 2.
A phylogenetic tree (UPGMA) inferred from the multiple alignments of amino acid sequences of histone-like proteins from dinoflagellate HCCs (a), bacterial histone-like protein family (b), and bacterial DNA-binding HU proteins (c) was constructed with PHYLIP version 3.5. The numbers at the nodes represent the bootstrap percentages by creation of 500 replicated data sets with SEQBOOT. The accession number for each protein sequence is as follows: HCC1 (C. cohnii), A56581; HCC2 (C. cohnii), B56581; BpH2 (B. pertussis), AAB40156; XHLP (X. fastidiosa), AAF84745; HU (Salmonella enterica serovar Typhimurium), S01732; HU (Streptococcus pneumoniae), AAK75224; HU (Haemophilus influenzae), P43722; HU (Pseudomonas aeruginosa), JC4061; HU (Deinococcus radiodurans), Q9RZ89.