Abstract
Partial nitrification of ammonium to nitrite under oxic conditions (nitritation) is a critical process for the effective use of alternative nitrogen removal technologies from wastewater. Here we investigated the conditions which promote establishment of a suitable microbial community for performing nitritation when starting from regular sewage sludge. Reactors were operated in duplicate under different conditions (pH, temperature, and dilution rate) and were fed with 50 mM ammonium either as synthetic medium or as sludge digester supernatant. In all cases, stable nitritation could be achieved within 10 to 20 days after inoculation. Quantitative in situ hybridization analysis with group-specific fluorescent rRNA-targeted oligonucleotides (FISH) in the different reactors showed that nitrite-oxidizing bacteria of the genus Nitrospira were only active directly after inoculation with sewage sludge (up to 4 days and detectable up to 10 days). As demonstrated by quantitative FISH and restriction fragment length polymorphism (RFLP) analyses of the amoA gene (encoding the active-site subunit of the ammonium monooxygenase), the community of ammonia-oxidizing bacteria changed within the first 15 to 20 days from a more diverse set of populations consisting of members of the Nitrosomonas communis and Nitrosomonas oligotropha sublineages and the Nitrosomonas europaea-Nitrosomonas eutropha subgroup in the inoculated sludge to a smaller subset in the reactors. Reactors operated at 30°C and pH 7.5 contained reproducibly homogeneous communities dominated by one amoA RFLP type from the N. europaea-N. eutropha group. Duplicate reactors at pH 7.0 developed into diverse communities and showed transient population changes even within the ammonia oxidizer community. Reactors at pH 7.5 and 25°C formed communities that were indistinguishable by the applied FISH probes but differing in amoA RFLP types. Communities in reactors fed with sludge digester supernatant exhibited a higher diversity and were constantly reinoculated with ammonium oxidizers from the supernatant. Therefore, such systems could be maintained at a higher dilution rate (0.75 day−1 compared to 0.2 day−1 for the synthetic wastewater reactors). Despite similar reactor performance with respect to chemical parameters, the underlying community structures were different, which may have an influence on stability during perturbations.
Strong regulations exist nowadays for the removal of ammonium from wastewater discharge. Ammonium can lead to eutrophication of freshwater bodies and is toxic to aquatic life (11). The current method of removal of ammonium in wastewater treatment plants consists of a combination of aerobic nitrification catalyzed by autotrophic organisms and anaerobic denitrification catalyzed by heterotrophic organisms (40). In a first step, ammonium is oxidized to nitrate via nitrite, and in a second step, nitrate is reduced with organic carbon via nitrite to N2. This combination was long considered as the only way to remove ammonium from wastewater. However, more recent economical considerations and discoveries urge either the use of shortcuts in the nitrification-denitrification process, by going via nitrite instead of nitrate (the Sharon process [13]), or application of the anammox process (12, 17, 38). In the anammox process ammonium is oxidized anaerobically with nitrite to N2 by autotrophic bacteria (18).
Although not directly applicable for the main stage in aerobic treatment, the combination of nitritation and denitrification (13) or nitritation and anammox (20, 34, 38, 39) has interesting economical advantages for the separated treatment of ammonium-rich wastewater, like rejection water from sludge digester tanks. In fact, the amount of ammonium derived from the sludge digester can represent more than 20% of the daily nitrogen load of the inlet of a wastewater treatment plant (33). A combination of partial nitrification and anammox would in the first place require less oxygen input for the nitrification part (since only a part of ammonium is oxidized to nitrite rather than all ammonium to nitrate). Secondly, no addition of external organic carbon is necessary, which is often practiced in the nitrification-denitrification scheme by the addition of methanol (13), since the anammox process is carried out by autotrophic bacteria. One possible configuration for a combination of nitritation and anammox would be a two-reactor system to control the different physiological requirements for both types of microorganisms (e.g., oxygen versus no oxygen). In the nitritation stage, the ammonia-oxidizing bacteria would produce the nitrite which is then used by the anammox bacteria. Since anammox bacteria require equimolar (or slightly higher) nitrite and ammonium loads (8, 16, 20, 31, 37, 39), the process would need to be balanced for ammonium depending on how much ammonium was converted by the ammonia oxidizers. Generation and maintenance of a nitritation reactor requires that nitrite-oxidizing bacteria are washed out from the biomass and can no longer find suitable conditions under which to reestablish themselves. Unfortunately, ammonia- and nitrite-oxidizing bacteria can be found almost everywhere, and therefore it might be difficult to find conditions favoring one over the other. Up to now, there are 25 cultured species of ammonia-oxidizing bacteria (21), all with different salt requirements and substrate affinities for ammonia and/or urea. Also, nitrite-oxidizing bacteria—of which there exist eight pure cultures (21)—differ in ecophysiological requirements. For example, members of Nitrospira generally are regarded as obligately chemolithotrophic, whereas members of Nitrobacter can also thrive on organic compounds for energy generation (3). Members of Nitrospira prefer relatively low nitrite concentrations (9) and are found as the most abundant nitrite oxidizer in wastewater treatment systems (5, 6, 21). Normal temperatures (5 to 20°C) and conditions in wastewater treatment plants favor growth of nitrite oxidizers, with the result that ammonium is completely oxidized to nitrate. Mathematical modeling with physiological parameters of Nitrosomonas and Nitrobacter was used to predict the success of nitritation (13, 14), and it was proposed that mesophilic temperatures (between 35 and 40°C) and pHs between 7 and 8 are required to compete more effectively than nitrite oxidizers. These and other studies concluded that the free NH3 concentration actually was the most important factor for selecting against nitrite oxidizers (2, 14). The competitive disadvantage of nitrite oxidizers to ammonia oxidizers at 35°C has been experimentally established in sludge reactors (13, 22). However, since Nitrobacter is not commonly found in wastewater treatment plants (5, 24), it was interesting to study if nitritation could also be established from activated sludge at mesophilic temperatures (25 and 30°C) and at near-neutral pHs (7 and 7.5).
Here we studied the effects of small temperature, pH, and dilution rate differences on the formation, composition, and activity of the ammonia-oxidizing biomass in bioreactors inoculated from sewage sludge. The ammonia-oxidizing community in the biomass of the nitritation reactors was quantitatively monitored over time by using fluorescence in situ hybridization (qFISH). Changes in the diversity of the ammonia oxidizers were further studied by determining restriction fragment length polymorphisms (RFLP) of amplified amoA gene fragments from community DNA. Furthermore, several different reactor configurations were independently repeated, in order to determine whether particular operating conditions would repeatedly lead to the same composition of ammonia oxidizers.
MATERIALS AND METHODS
Nitritation reactor setup and growth conditions.
Nitritation reactors were inoculated from regular activated sludge and operated for periods of up to 50 days. The influences of pH (7 or 7.5), dilution rate (from 0.2 up to 1 day −1), temperature (25 or 30°C) and medium (synthetic [see below] or sludge digester supernatant) on the community development, nitrite and nitrate production, and ammonium removal were tested (for a detailed operation plan, see Table 1). All sludge experiments were performed in pH- and temperature-controlled bioreactors (Bioengineering, Wald, Switzerland) in a total volume of 2.5 to 3 liters. Each reactor was inoculated with activated sludge from the aeration basin from Werdhölzli, the wastewater treatment plant of Zürich, Switzerland, which treats approximately 200,000 m3 of wastewater each day. For inoculation, sludge biomass (same volume as used for the reactors) was immediately transported from the treatment plant to the laboratory, centrifuged for 10 min at 8,000 rpm (Centrikon H403 centrifuge; Kontron Instruments, Volketswil, Switzerland), and resuspended in 2.5 to 3 liters of synthetic mineral medium (depending on the reactor volume) or in the same amount of sludge digester supernatant. Synthetic mineral medium contained 50 mM NH4HCO3, 50 mM phosphate buffer (as potassium salt), 10 mM KHCO3, 3 mM Na2SO4, 0.5 mM CaCl2, 0.5 mM MgCl2, and 1 mM EDTA in demineralized water. Furthermore, 2 ml of trace element solution 1 (containing 10 g of Na2-EDTA · 2H2O and 5 g of FeSO4 per liter of demineralized water) and 1 ml of trace element solution 2 (containing, per liter of demineralized water, 15 g of Na2-EDTA · 2H2O, 0.43 g of ZnSO4 · 7H2O, 0.24 g of CoCl2 · 6H2O, 0.99 g of MnCl2 · 4H2O, 0.25 g of CuSO4 · 5H2O, 0.22 g of NaMoO4 · 2H2O, 0.19 g of NiCl2 · 6H2O, 0.08 g of Na2SeO3, and 0.014 g of H3BO4) were added per liter of mineral medium. Sludge digester supernatant was obtained from the sludge digesting tanks at Werdhölzli and immediately used as feed for the respective reactors (Table 1). The approximate salt concentrations of the sludge supernatant were as follows: chloride, 1.9 mM; sulfate, 0.08 mM; phosphate, <50 μM; Na+, 3.9 mM; K+, 3.1 mM; Mg2+, 0.9 mM; Ca2+, 2.6 mM; NO3−, <50 μM; NH4+, 56 mM.
TABLE 1.
Overview of the different reactor operating conditions
| Reactor | Conditions | Figure | Starting biomass | Medium | Inoculation | Remarks |
|---|---|---|---|---|---|---|
| 1A | pH 7.5, 30°C | 2-4A | Activated sludge | Synthetic | Single | Dilution rate from 0.2 to 0.3 day−1 at day 25; half amount of starting sludge |
| 1B | Repetition of 1A | 2-4B | Activated sludge | Synthetic | 1B and 2B | pH increase at day 19 from 7.5 to 7.9 |
| 2A | pH 7, 30°C | 2-4C | Activated sludge | Synthetic | 2A and 3A | |
| 2B | Repetition of 2A | 2-4D | Activated sludge | Synthetic | 2B and 1B | pH decrease at day 30 from 7 to 6.6 |
| 3A | pH 7.5, 25°C | 2-4E | Activated sludge | Synthetic | 3A and 2A | |
| 3B | Repetition of 3A | 2-4F | Activated sludge | Synthetic | 3B and 5 | |
| 4 | pH 7, 30°C | 2-4G | Activated sludge and supernatant of sludge digester | Supernatant of sludge digester | Single | Dilution rates (day−1): 0.2 (day 0-15), 0.3 (day 15-17), 0.5 (day 17-20),0.75 (day 20-22), 1.0 (day 22-27) |
| 5 | pH 7, 25°C | 2-4H | Activated sludge and supernatant of sludge digester | Supernatant of sludge digester | 5 and 3B |
Reactor cultures were flushed with a mixture of oxygen and CO2 to prevent an increase of the pH. For the experiments at pH 7.5, the cultures were flushed with Aligal28 (20% CO2 and 80% O2 with at least 99.5% purity). Reactors operated at pH 7 were flushed with a gaseous mixture of 30% CO2 (99.99% purity) and 70% O2 (99.95%). All gases were purchased from Carbagas (Rümlang, Switzerland). The impeller speed of the reactor was 500 rpm, and the pH was further maintained by automatic addition of 2 M KOH when necessary.
Nucleic acid extractions.
Total genomic DNA was isolated from sludge samples from the different reactors at regular time intervals. Biomass (of up to 50 ml of sludge) was pelleted by centrifugation and resuspended in 0.9 ml of TEN homogenization buffer (TEN is a solution of 0.1 M NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH 8.0) in a screw-cap vial. Glass beads (0.2 g; 0.1 mm in diameter) were added, and the mixture was shaken twice for 1 min at 4,000 rpm in a Braun cell homogenizer (Inotech AG, Dottikon, Switzerland) with a 1-min interval on ice. The glass beads were allowed to settle without centrifugation, and the supernatant was transferred to a fresh Eppendorf tube. This sample was mixed 1:1 with Tris-buffered phenol (pH 8). After vortexing, 1 volume of chloroform-isoamylalcohol (24:1 vol/vol) was added. After vortexing and centrifugation for 10 min at 15,000 × g and 4°C, the water phase was transferred to a new Eppendorf tube and mixed with the same volume of chloroform-isoamyl alcohol (24:1, vol/vol), vortexed, and centrifuged for 10 min as before. DNA was precipitated with 2 volumes of ethanol and 0.1 volume of 3 M sodium-acetate (pH 5.5) for 1 h at −80°C or overnight at −20°C and recovered by centrifugation. The pelleted nucleic acids were washed with a solution of 70% (vol/vol) ethanol plus 30% TE (TE is 10 mM Tris-HCl plus 1 mM EDTA, pH 8) and dissolved in 10 mM Tris-HCl, pH 7.5.
Amplification of amoA.
Fragments of 491 bp of the amoA gene coding for the active subunit of ammonium monooxygenase were amplified from isolated sludge community DNA by PCR. Primers for amoA amplification were those published previously: amoA-1F (5′-GGGGTTTCTACTGGTGGT-3′) and amoA-2R [5′-CCCCTC(G/T)G(G/C)AAAGCCTTCTTC-3′] (29). PCR reagents were used according to the supplier (Sigma, Buchs, Switzerland). PCR with amoA primers was carried out under the following cycling regime: 4 min at 95°C; 35 cycles each of 1 min at 95°C and 45 s 60°C; and 1 min 72°C. Final extension was carried out for 3 min at 72°C. The amoA PCR-products were either used directly for RFLP studies (see below) or cloned in vector pGEM-T-Easy (Promega, Wallisellen, Switzerland) and transformed into Escherichia coli DH5α by established procedures (30). Selected amoA inserts were sequenced on GenElute-isolated plasmid DNA (Sigma) on both strands by using the Thermosequenase kit (Amersham Biosciences Europe, Dübendorf, Switzerland) with IRD-800- and IRD-700-labeled universal vector-located primers (MWG Biotech, Ebersberg, Germany). Sequence transcripts were separated and analyzed on a LiCOR (Lincoln, Nebr.) 4200L IR2 automated DNA sequencer.
RFLP.
Amplified amoA fragments from community DNA of the reactors at different time points were digested simultaneously with RsaI and HaeIII (Amersham Biosciences Europe). Digested products were separated on SPREADEX EL 400 precast gels (Elchrom Scientific AG, Cham, Switzerland) together with a DNA size marker (M3; Elchrom), with a separation range from approximately 20 to 400 bp. Gel electrophoresis was performed on a SEA 2000 apparatus (Elchrom) during 81 min (with buffer circulation and constant temperature of 55°C) to ensure optimal and reproducible separation of the restriction fragments. After electrophoresis, the DNA was stained with SYBR gold (0.05 μl of concentrated stock/ml; Molecular Probes Europe BV, Leiden, The Netherlands) and visualized under UV light. Digital images were recorded with the Q-EL 330 digital gel recording and analysis system (Elchrom).
FISH.
Cells from up to 50 ml of sludge culture were recovered by centrifugation and resuspended in phosphate-buffered saline (PBS), pH 7.4, consisting of 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 per liter of distilled water. The samples were fixed with 4% paraformaldehyde (in PBS) for 1 h at room temperature. Afterwards, the cells were washed twice with PBS. The fixed cells were suspended in a solution of 50% PBS and 50% ethanol and stored at −20°C. For FISH, 10 μl of the fixed sample (or multiple 10-μl aliquots for qFISH) was applied on a well of a gelatin-coated (1) glass slide (Huber & Co., Reinach, Switzerland), dried for 1 h at 46°C, and subsequently dehydrated in solutions of 50%, 80% (vol/vol, in 10 mM Tris-HCl, pH 7.5), and 96% ethanol for 3 min each. To start hybridization, 9 μl of hybridization buffer (with a composition dependent on the used probe) (Table 2) and 1 μl of fluorescently labeled probe (at a concentration of 50 ng/μl) were added to a well. If necessary an unlabeled competitor (at 50 ng/μl) was added to the mixture (Table 2). The hybridization was conducted for 1 h at 46°C in a humidified chamber.
TABLE 2.
Probes used for FISH and the corresponding hybridization and washing conditions
| Probe | Sequence (5′→3′) | Target organisms | Conca
|
Competitor | Reference | |
|---|---|---|---|---|---|---|
| Formamide (%) | NaCl (M) | |||||
| EUB338 | GCTGCCTCCCGTAGGAGT | Bacteria | 0 | 0.9 | 1 | |
| EUB338II | GCAGCCACCCGTAGGTGT | Bacteria | 0 | 0.9 | 4 | |
| EUB338III | GCTGCCACCCGTAGGTGT | Bacteria | 0 | 0.9 | 4 | |
| NEU | CCCCTCTGCTGCACTCTA | Halophilic and halotolerant members of the genus Nitrosomonas | 40 | 0.056 | CTE | 41 |
| Ntspa662 | GGAATTCCGCGCTCCTCT | Nitrospira genus | 35 | 0.08 | CNtspa662 | 5 |
| NmII | TTAAGACACGTTCCGATGTA | Many members of the Nitrosomonas communis sublineage | 25 | 0.159 | 26 | |
| Nsv443 | CCGTGACCGTTTCGTTCCG | Most Nitrosospira spp. | 30 | 0.112 | 23 | |
| Nso1225 | CGCCATTGTATTACGTGTGA | Almost all ammonia-oxidizing β-Proteobacteria | 35 | 0.08 | 23 | |
| Nso190 | CGATCCCCTGCTTTTCTCC | Many but not all ammonia-oxidizing β-Proteobacteria | 50 | 0.028 | 23 | |
| Nmo218 | CGGCCGCTCCAAAAGCAT | Many members of the Nitrosomonas oligotropha sublineage | 35 | 0.08 | 10 | |
| NmV | TCCTCAGAGACTACGCGG | Nitrosococcus mobilis | 35 | 0.08 | 26 | |
| Nit3 | CCTGTGCTCCATGCTCCG | Nitrobacter | 40 | 0.056 | CNIT3 | 42 |
| CNIT3 | CCTGTGCTCCAGGCTCCG | Competitor to Nit3 | 42 | |||
| CTE | TTCCATCCCCCTCTGCCG | Competitor to NEU | ||||
| CNtspa662 | GGAATTCCGCTCTCCTCT | Competitor to Ntspa662 | ||||
Concentrations presented as percentage of formamide in hybridization buffer or molar concentration of NaCl in wash buffer.
Following hybridization, a stringent washing step was performed for 10 min at 48°C in a buffer with the appropriate NaCl concentration (Table 2). Samples were counterstained with 4,6-diamidino-2-phenylindole (DAPI) at a final concentration of 10 mg/liter for 5 min at room temperature (15) and mounted in Citifluor (Citifluor, London, United Kingdom). All oligonucleotide probes were obtained from Microsynth (Balgach, Switzerland). Microscopy was performed on an Olympus BX50 microscope, equipped with filters HQ-CY3 and HQ-DAPI (both from Analysentechnik AG, Tübingen, Germany). Digital images were taken with a Sensys cooled charge-coupled device camera (Photometrics, Tucson, Ariz.) and acquired in the program METAVIEW (Visitron, Puchheim, Germany).
Quantative FISH was performed with a Zeiss (Jena, Germany) CSLM 510 confocal laser scanning microscope equipped with a HeNe laser at 543 nm and an Ar ion laser at 488 nm. Population sizes were determined as the relative biovolume taken up by cells stained with the Cy3-labeled specific probe compared to the biovolume of cells stained with all three Fluos-labeled EUB probes (7, 31).
Analytical measurements.
Concentrations of nitrite, nitrate, chloride, sulfate, and phosphate were determined by ion-exchange chromatography using a DX500 apparatus with an IONPAC-ATC1 anion trap column, a IONPAC-AG11 guard column, an analytical IONPAC-AS11 4-mm column, an ASRSII 4-mm suppressor, and a CD20 conductivity detector (from Dionex, Olten, Switzerland). The concentrations of ammonium, sodium, potassium, magnesium, and calcium were quantified by ion-exchange chromatography using the same DX500 apparatus but with an IONPAC-CTC1 cation trap column, a IONPAC-CG12 guard column, an analytical IONPAC-CS12 4-mm column, a CSRS Ultra 4-mm suppressor, and a CD20 conductivity detector (Dionex, Olten, Switzerland).
Protein concentrations in sludge samples were measured using the Bio-Rad protein assay (Bio-Rad Laboratories, GmbH, Munich, Germany). Cells were pelleted, resuspended in 0.1 M NaOH, and incubated for 1.5 h in a boiling water bath. An aliquot (0.8 ml) of the diluted suspension was mixed with 0.2 ml of Bio-Rad dye reagent and incubated for 15 min in the dark. The absorbance was measured at 595 nm. A standard curve was created using known concentrations of bovine serum albumin fraction V (Sigma).
RESULTS
Nitritation activity under synthetic medium conditions.
The effects of pH and temperature on the formation of active ammonia-oxidizing cultures and resulting in the absence of nitrite-oxidizing cultures were tested in continuously stirred tank reactors first with sterile synthetic medium containing 50 mM ammonium and HCO3− as the sole carbon source. In all cases nitrite production started 5 to 10 days after inoculating the reactors with fresh activated sludge, increasing afterwards to reaching effluent concentrations of between 30 and 50 mM nitrite, except for reactor 3B, where a slightly higher ammonium concentration was fed (Fig. 1). Under all conditions, except in reactor 3A, the ammonium concentration dropped from an influent concentration of 50 mM to levels around 10 mM or less. This indicated that nitritation was effectively taking place. Nitrite-oxidizing activity was small in most reactors and limited to the first five days after inoculation. Afterwards, a slow decline was observed. Nitrate was never produced at concentrations higher than 7 mM. Protein concentrations—an indication for the total biomass in the reactors—decreased constantly from between 200 and 300 μg of protein/ml directly after inoculation to more or less constant levels after 2 weeks of between 20 and 30 μg of protein/ml (not shown). This was expected, since the community was forced to change from mixed heterotrophic (in activated sludge) to one dominated by autotrophic organisms (due to carbonate being the only carbon source in the medium).
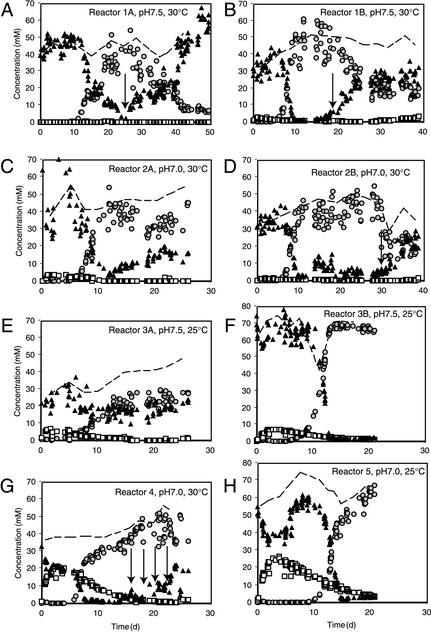
FIG. 1.
Chemical nitrogen parameters in the different reactors during incubation time. (A) Reactor 1A; (B) reactor 1B; (C) reactor 2A; (D) reactor 2B; (E) reactor 3A; (F) reactor 3B; (G) reactor 4; (H) reactor 5. Panels correspond to Table 1 and to those in Fig. 2 and 3. Shown are concentrations of ammonium in the effluent (black triangles), nitrate (open squares), nitrite (grey circles), and total nitrogen (dotted line) (as the sum of ammonium, nitrate, and nitrite concentrations). Since total nitrogen was solely determined arithmetically, any measuring errors in its components (i.e., ammonium, nitrite, and nitrate) contribute to unrealistic fluctuations. Note the different time scales for the experiments in reactors 1A, 1B, and 2B. Arrows point to important changes in operating parameters, as specified in Table 1.
Contrary to our expectations, no large differences occurred in soluble nitrogen parameters (ammonium, nitrite, and nitrate) over time at different pHs and temperatures. The only consistent observation was a stronger nitrate production at lower incubation temperatures and a slightly longer lag in the occurrence of nitrite production (Fig. 1E and F). Due to technical reasons, not all reactors could be operated at the same time, but repetitions of the nitritation reactors at different starting points (and therefore presumably slightly different composition of inoculated sludge) resulted in mostly similar chemical process parameters (Fig. 1C and D).
Nitritation activity with sludge digester supernatant.
Two reactors were operated with sludge digester supernatant instead of synthetic medium (reactors 4 and 5). In those two cases, nitrite production also started after about 10 days, but relatively high amounts of nitrate (20 to 25 mM) were produced during the first 2 weeks of operating time (Fig. 1G and H). After 5 days, however, no new net nitrate production occurred and nitrate washed out from both reactors according to the dilution rate (not shown). The peak and subsequent decline in nitrate production occurred before the onset of measurable nitrite concentrations in the effluent. Nitrite production started earlier in the reactor operated at 30°C than the one at 25°C, which is consistent with the observations for the reactors running with synthetic medium. However operating conditions, both reactors finally established effective ammonium oxidation with high nitrite but low nitrate concentrations.
Effects of dilution rates and pH stability.
In two cases, the effect of increasing dilution rate on nitrite production was tested. In reactor 1A (operated with synthetic medium at pH 7.5 and 30°C) an increase of dilution rate from 0.2 to 0.3 day−1 from day 25 to day 50 resulted in a complete washout of ammonia-oxidizing activity from the reactor (Fig. 1A). Ammonium concentrations after day 45 reached 50 mM, and nitrite concentrations in the effluent decreased to less than 5 mM. Reactor 4, which was fed with sludge digester supernatant, showed a completely different behavior (Fig. 1G). In this case, the dilution rate could be increased to about 0.75 day−1 without dramatic increase of ammonium levels in the effluent. Only at a dilution rate of 1.0 day−1 (after day 22) did ammonium concentrations start to increase above 10 mM in the effluent. This indicated that reactors fed with sludge digester supernatant can be operated at higher dilution rates. This difference seemed to be at least partly the result of a reinoculation of ammonia oxidizers from the nonsterile sludge digester supernatant (see below).
The effects of pH changes on the activity of a stable nitritation system were investigated in two reactors. A pH change from 7.5 to 7.9 in reactor 1B at day 19 (operated with synthetic medium at 30°C) resulted in an immediate loss of ammonia-oxidizing activity. Ammonium concentrations in the effluent increased and nitrite concentrations decreased to about 20 mM (Fig. 1B). A decrease from pH 7 to pH 6.6 in reactor 2B at day 30 and beyond also resulted in partial loss of ammonia-oxidizing activity. Both nitrite and ammonium concentrations in the effluent changed to between 20 and 25 mM (Fig. 1D). These results indicated that the nitritation systems were relatively sensitive to pH changes outside the optimal range of 7 to 7.5.
Population analysis of the communities in different reactor configurations.
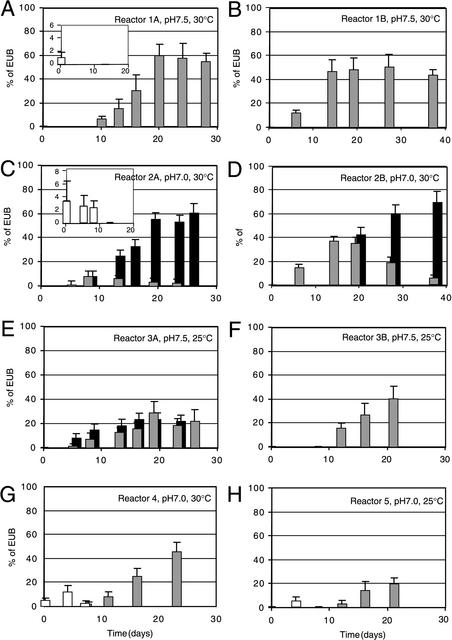
We were interested to determine whether specific operating conditions would lead to very defined communities of ammonia oxidizers in the reactors. Thus, sludge samples were analyzed at regular time intervals by qFISH and amoA RFLP. According to our focus we only applied FISH-16S rRNA probes targeting the ammonia and nitrite-oxidizing community (Table 2). At the time of inoculation the activated sludge contained relatively low numbers of ammonia-oxidizing bacteria. Members of the Nitrosomonas oligotropha sublineage (represented by Nmo218 staining) were present in 4.5% ± 3.5% of EUB-stainable cells (all percentages are means ± standard deviations), whereas the presence of Nitrosomonas communis members (NmII stainable) was 4% ± 3% and that of Nitrosomonas stainable with NEU was below 1%; no ammonia-oxidizing bacteria stainable with Nsv443 or NmV were present. Nitrite-oxidizing bacteria from the genus Nitrospira (judged from cells stainable with Ntspa662) were present in the inoculated sludge at relative levels of 3% ± 2.5%. No bacteria stainable with the Nitrobacter probe (Nit3) were detected.
After starting the nitritation reactors, strong shifts in community composition of ammonia and nitrite oxidizer populations were observed. In general, these shifts followed the trends observed from chemical analysis of nitrogen parameters (Fig. 1). The community of ammonia-oxidizing bacteria increased under all conditions, parallel to the observed increase of nitrite concentrations in the effluent. On the other hand, the nitrite oxidizer community never established itself but decreased and was no longer quantifiable after 10 days (Fig. 2A, C, and G). Interestingly, similar nitritation activities (Fig. 1) were found with different ammonia-oxidizing communities. Under conditions of pH 7.5 and 30°C and with synthetic medium, the community of ammonia oxidizers developed to an almost exclusive population of NEU-stainable cells (Fig. 2A and B). In the biomass of these two reactors, the relative amount of cells stained with the more general probe Nso190 was the same as that stained with NEU. Both reactors operated at pH 7 and 30°C established different groups of ammonia-oxidizing bacteria. In both reactors, there was an initial increase of a population stainable with the NEU probe (Fig. 2C and D), which decreased after approximately 10 days of incubation at the expense of an increase of a second group of ammonia oxidizers, stainable with the more general probe Nso190. Under conditions of pH 7.5 and at 25°C the increase in the population of ammonia oxidizers was less pronounced than at 30°C (Fig. 2E and F). FISH community quantification with the probes NEU and the more general probe Nso190 gave the same values, suggesting that the ammonia-oxidizing community under these conditions again developed reproducibly to one dominated by NEU-stainable cells. This pointed to pH being more important than temperature in selecting ammonia oxidizers stainable with NEU.
FIG.2.
Community composition development of ammonia and nitrite oxidizers in the different reactors during operation time. Panels correspond to those in Fig. 1 and 3. Shown are relative biovolumes of FISH probe-stainable groups compared to the biovolume taken up by the biomass stainable with the EUB probe mixture (% of EUB). Probes used for quantitation were NEU (grey bars), which targets several halophilic and halotolerant members of the genus Nitrosomonas; Nso190 (black bars), representative for many but not all ammonia-oxidizing bacteria from the β-subgroup of Proteobacteria; and Ntspa662 (white bars), specific for all members of the genus Nitrospira. Since we observed that the fluorescence intensities obtained with Nso1225 were weaker than those obtained with Nso190, we decided not to use Nso1225 for quantitative FISH. When visual inspection indicated that all aggregates were stained with NEU, no specific FISH with Nso190 was performed (as in panels A, B, F, G, and H), except for reactor 3A, which was used to confirm that in such a case NEU- and Nso190-based counts were the same. Note that in some instances the population sizes were detectable but appear as very small bars on the scale (e.g., days 0, 6, and 8 in some reactors). The inset in panels A and C shows the Nitrospira populations on an enlarged scale. Note the different time axes in panels B and D.
Both reactors operated with sludge digester supernatant also developed to a community dominated by NEU-stainable cells. Despite a pH of 7.0, no transition to other Nso190-stainable organisms took place in these reactors, since relative levels of the Nso190-stainable population were the same as those of the NEU-stainable population and the probe NEU stained all nitrifying aggregates (not shown). It might be that in the case of these two reactors the increased dilution rate or the salt composition of the sludge digester supernatant were selecting for the NEU-population.
Diversity of the amoA gene in different communities.
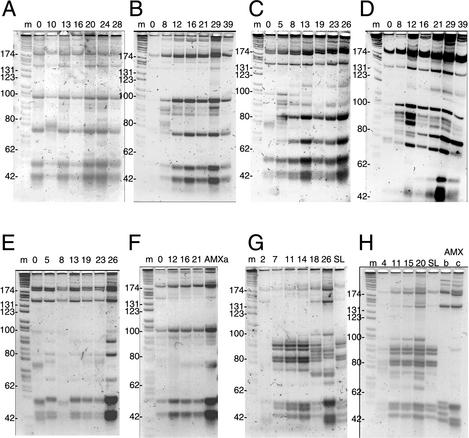
Since most FISH probes applied in this study were not species specific, we used RFLP analysis of the amoA gene to investigate diversity of the involved ammonia-oxidizing bacteria under the different reactor conditions at a higher resolution. For this purpose, sludge DNA was extracted, a part of the amoA gene was amplified by PCR and digested with two enzymes (RsaI and HaeIII), and the products were separated on high-resolution acrylamide gel (Fig. 3). RFLP patterns of amoA amplified DNA from both reactors operated at pH 7.5 and 30°C with synthetic medium were highly similar during the entire incubation period (Fig. 3A and B), except for some minor banding differences. The amoA RFLP of this population was mostly represented by the banding pattern derived from one of the cloned sequences (see below, Fig. 3F, lane of amoA clone AMXa). As expected from qFISH analyses, the RFLP patterns in reactors running under conditions of pH 7.0 and 30°C differed considerably from those at pH 7.5 and 30°C and at pH 7.5 and 25°C. Moreover, both reactors operated at pH 7.0 and 30°C developed different ammonia oxidizer communities in time (Fig. 3C and D). Reactor 2A (Fig. 3C) showed a transient occurrence of the same AMXa RFLP group as observed in reactors 1A and 1B between days 0 and 13, which was then taken over by a different RFLP group. Representative bands for the AMXa group of reactor 1 (183, 97, 71, 52, 41, and 40 bp) were also visible in reactor 2B (Fig. 3D) and more intense as in reactor 2A. This is consistent with the changes in relative amounts of NEU-stainable cells (Fig. 2D). However, from day 16 onwards a different RFLP pattern started to appear (represented by a double band at 185 bp and bands at 140, 79, and 66 bp). These RFLP patterns originate from different amoA sequences. At pH 7.5 and 25°C again a different type of community developed, although this had not been observed by qFISH analysis (Fig. 3E and F). In addition, the communities in both these reactors were not identical. In reactor 3A a dominant community developed which was represented by bands at 140, 50, 45, and 42 bp (Fig. 3E). This was most similar to the RFLP type of cloned sequence AMXc (Fig. 3H, lane AMXc). The community in reactor 3B was dominated by an RFLP type similar to that observed in reactors 1A and 1B, although consistently the 79-bp band was lacking (Fig. 3F). Both communities were relatively homogenous at all time points, indicating that no major transitions in the community structure took place. Patterns in the communities of reactors 4 and 5, which were fed with supernatant of sludge digester, were very similar to one another but very different from those of the reactors fed with synthetic medium (Fig. 3G and H). Interestingly, the RFLP patterns derived from the community of the reactors were also very similar to that of DNA from the biomass of the sludge digester supernatant itself (Fig. 3G and H, lanes SL). This was a confirmation for the hypothesis that those reactors were continuously inoculated from the nonsterile sludge digester feed. We suspect that different populations make up the community of ammonia oxidizers in reactors 4 and 5. For example, the pattern of cloned sequence AMXb (Fig. 3H, lane AMXb) could be found among the complex patterns in reactors 4 and 5. The effect of increasing dilution rate in reactor 4 was also immediately visible as a change in community structure (Fig. 3G, lanes 18 and 26), which had not become evident from FISH analysis.
FIG. 3.
RFLP patterns of the amoA amplifiable DNA in the communities from the different reactors. Panels correspond to the assignments given in Fig. 1 and 2. Sizes of some of the bands in the M3 marker are indicated in base pairs to the left of each panel. Lanes are marked according to the sampling day or as M3 marker (m); as AMXa, AMXb, and AMXc, cloned and sequenced amoA fragments; or as SL, incoming biomass from the sludge digester.
From each of the reactors 1A, 2A, and 3A, approximately 10 cloned amoA gene fragments were sequenced. From reactor 1A, 1 clone was from DNA isolated at day 28 and 9 clones of DNA were retrieved after 31 days of operation, from reactor 2A clones were found in DNA from days 8 (1 clone) and 19 (9 clones), and from reactor 3A clones were found in DNA from days 13 (1 clone) and 19 (10 clones). One type of sequence was detected among the reactor 1A clones, (named AMXa) which had the highest homology to GenBank entry AF272483 (98%). This sequence groups in the Nitrosomonas europaea-Nitrosomonas eutropha sublineage. The RFLP pattern of this cloned sequence (Fig. 3F, lane AMXa) was indeed representative for the patterns in reactors 1A and B. All sequences from the reactor 2A and eight from reactor 3A (one from day 13 and 7 at day 19) displayed 99% nucleotide identity to GenBank entry AF276485 (named AMXc). This sequence groups in the Nitrosomonas cluster 6a containing the cultured species N. urea and N. oligotropha (28). The RFLP type of AMXc was found in reactors 2A and 3A (Fig. 3C and E). One additional sequence cloned from reactor 3A DNA (AMXb, present in three clones at day 19) displayed 99.8% identity to the sequence in AF276476. This sequence grouped near cluster 6 (28); however, its RFLP type was not clearly visible in reactor 3A, but only in the sludge digester supernatant for reactor 4 (Fig. 3G).
DISCUSSION
Despite only small differences in operating parameters (pH 7.0 or 7.5; temperature of 25 or 30°C), the communities of ammonia oxidizers which developed in the nitritation reactors were remarkably different. Only at pH 7.5 and 30°C the sludges developed into mostly identical communities of ammonia oxidizers belonging to the N. eutropha-N. europaea subgroup (stainable with probe NEU and represented by amoA clone sequence AMXa), even though the reactors were started at different time points with different sludges. All other conditions led to communities either stainable with the same FISH-probe but different in amoA RFLP (at 25°C and pH 7.5), or even groups stainable with different FISH probes (at pH 7.0 and 30°C). This suggests that pH is more important than temperature in selecting N. eutropha-N. europaea ammonia oxidizers. High ammonium loads were also found to be a selective parameter for growth of N. eutropha, but only occurring at concentrations above 65 mM ammonium (27), which was not varied in our experiments. The selection of ammonia oxidizers from the subgroup of N. eutropha-N. europaea was also reported by others for nitritation experiments carried out under similar pH (7.2 to 8), temperature (30 to 35°C) and ammonium loads (22, 35, 38), but to our knowledge, never before were community shifts of ammonia oxidizers in otherwise complex systems followed in such detail as described here. The two reactors operated with sludge digester supernatant (reactors 4 and 5) established a community with very similar RFLP patterns, at least until the time point of changing dilution rates (reactor 4, day 18) even though the reactor temperatures were different. The composition of this community was clearly determined by continuous reinoculation from the sludge digester supernatant, which may therefore be an important factor in technical scale applications. Since many studies looked at the performance of nitrification reactors fed with synthetic wastewaters (24, 25, 32, 35, 38) important differences to the community composition and development in technical systems which are subject to continuous reinoculation may have been overlooked. Furthermore, most studies, unfortunately, did not repeat reactor experiments independently, which therefore may have led to generalized conclusions on the composition and behavior of ammonia oxidizer communities where such are not to be made.
One important conclusion we drew from our work is that communities of ammonia oxidizers with different composition and complexity may form in reactor systems without obvious differences in performance of chemical parameters (such as rates of nitrite formation or ammonium oxidation). From a perspective of performance stability, it might be wiser to chose conditions favoring a more complex community of ammonia oxidizers. However, it was not tested systematically in our experiments whether more complex communities of ammonia oxidizers were indeed more resistant to variations in operating conditions. Both the relatively homogeneous sparse communities at pH 7.5 and 30°C (reactors 1A and B) were easily disturbed by a slight increase in dilution rate and pH. On the other hand, also the more diverse community at pH 7.0 and 30°C (reactor 2B), lost part of its nitritation activity soon after a pH decrease. The only system resisting a strong dilution rate increase was the complex community established with the sludge digester supernatant at pH 7.0 and 30°C (reactor 4), although with an equally strong shift in community composition.
Surprisingly, all conditions tested led to a development of sludges performing nitritation without nitrate formation, at least for the longevity of the experiment. Nitrite-oxidizing bacteria from the genus Nitrospira were present in the beginning of all reactor incubations, but gradually lost activity and were washed out almost completely from the reactors. This could be demonstrated clearly by qFISH. The specific conditions which supposedly promote preferential growth of ammonia-oxidizing bacteria over nitrite oxidizers have been intensively studied, although they still remain somewhat obscure, since not all ammonia and nitrite oxidizers catalyzing nitrification in wastewater treatment plants are available as pure culture (6, 19) and ammonia and nitrite oxidizers can develop under almost any environmental condition. Nitrite oxidizers in our experiments definitively remained more and longer active at lower temperatures (25°C) and in the reactors operated with sludge digester supernatant, which is in agreement with results from others who had shown that lower temperatures generally favor nitrite-oxidizing bacteria over ammonia-oxidizing bacteria (2, 13). On the other hand, the temperature difference alone cannot explain the better survival of nitrite oxidizers in the reactors operated with sludge digester rejection water. Nitrate production stopped very soon after inoculation in all reactor systems (after approximately 5 days), already before nitrite started to accumulate in the reactor. This suggests that perhaps the high ammonium concentrations prevailing at that time in the reactor (around 50 mM) were also responsible for the decline of nitrite-oxidizing activity. This would be in agreement to predictions that the amount of free ammonia is also responsible for inactivating nitrite-oxidizing bacteria (2, 13). Ammonia toxicity was reported to start at NH3 concentrations above 4 mg/liter (2, 13), which (at the 50 mM NH4+ in our experiments) at pH 7.0 and 7.5 (and 30°C) equaled approximately 9 and 20 mg/liter, respectively.
In conclusion, our results showed that stable nitritation can be achieved relatively easily by starting from activated sludge under a variety of conditions, although the underlying community structures of ammonia oxidizers may actually be rather different. The consequences of establishing different community structures for process stability are still poorly known but are worth studying more intensively, provided that community structures are reproducibly analyzed in sufficient detail to allow good comparisons to be drawn.
Acknowledgments
The work of K.E. was funded by a grant from BUWAL (Swiss Agency for the Environment, Forests, and Landscape); the Cantons of Luzern, St. Gallen, and Zürich; the Buholz (Luzern) and Werhölzli (Zürich) wastewater treatment plants; and the Office for Waste Disposal St. Gallen. M.W. was supported by a grant of the DFG (WA 1558/1-1).
We thank Kilian Stöcker for help with the confocal laser scanning microscopy.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, W., S. Baek, J. Chung, and Y. Lee. 2001. Optimal operational factors for nitrite accumulation in batch reactors. Biodegradation 12:359-366. [DOI] [PubMed] [Google Scholar]
- 3.Bock, E. 1976. Growth of Nitrobacter in the presence of organic matter. Arch. Microbiol. 108:305-312. [DOI] [PubMed] [Google Scholar]
- 4.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe eub338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 5.Daims, H., P. H. Nielsen, J. L. Nielsen, S. Juretschko, and M. Wagner. 2000. Novel Nitrospira-like bacteria as dominant nitrite-oxidizers in biofilms from wastewater treatment plants: diversity and in situ physiology. Water Sci. Technol. 41:85-90. [Google Scholar]
- 6.Daims, H., J. L. Nielsen, P. H. Nielsen, K. H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daims, H., N. B. Ramsing, K. H. Schleifer, and M. Wagner. 2001. Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl. Environ. Microbiol. 67:5810-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egli, K., U. Fanger, P. J. J. Alvarez, H. Siegrist, J. R. van der Meer, and A. J. B. Zehnder. 2001. Enrichment and characterization of an anammox bacterium from a rotating biological contacter treating ammonium-rich leachate. Arch. Microbiol. 175:198-207. [DOI] [PubMed] [Google Scholar]
- 9.Ehrich, S., D. Behrens, E. Lebedeva, W. Ludwig, and E. Bock. 1995. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium. Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch. Microbiol. 164:16-23. [DOI] [PubMed] [Google Scholar]
- 10.Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagopian, D. S., and J. G. Riley. 1998. A closer look at the bacteriology of nitrification. Aquacult. Eng. 18:223-244. [Google Scholar]
- 12.Hao, X., J. J. Heijnen, and M. C. M. van Loosdrecht. 2002. Sensitivity analysis of a biofilm model describing a one-stage completely autotrophic nitrogen removal (CANON) process. Biotechnol. Bioeng. 77:266-277. [DOI] [PubMed] [Google Scholar]
- 13.Hellinga, C., A. A. J. C. Schellen, J. W. Mulder, M. C. M. van Loosdrecht, and J. J. Heijnen. 1998. The Sharon process: an innovative method for nitrogen removal from ammonium-rich waste water. Water Sci. Technol. 37:135-142. [Google Scholar]
- 14.Hellinga, C., M. C. M. van Loosdrecht, and J. J. Heijnen. 1999. Model based design of a novel process for nitrogen removal from wastewater. Math. Comp. Model Dyn. 5:351-371. [Google Scholar]
- 15.Hicks, R. E., R. I. Amann, and D. A. Stahl. 1992. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl. Environ. Microbiol. 58:2158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hippen, A., C. Helmer, S. Kunst, K. H. Rosenwinkel, and C. F. Seyfried. 2001. Six years' practical experience with aerobic/anoxic deammonification in biofilm systems. Water Sci. Technol. 44:39-48. [PubMed] [Google Scholar]
- 17.Jetten, M. S., S. Logemann, G. Muyzer, L. A. Robertson, S. de Vries, M. C. van Loosdrecht, and J. G. Kuenen. 1997. Novel principles in the microbial conversion of nitrogen compounds. Antonie Leeuwenhoek Int. J. 71:75-93. [DOI] [PubMed] [Google Scholar]
- 18.Jetten, M. S., M. Strous, K. T. van de Pas-Schoonen, J. Schalk, U. G. van Dongen, A. A. van de Graaf, S. Logemann, G. Muyzer, M. C. van Loosdrecht, and J. G. Kuenen. 1998. The anaerobic oxidation of ammonium. FEMS Microbiol. Rev. 22:421-437. [DOI] [PubMed] [Google Scholar]
- 19.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Röser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch, G., K. Egli, J. R. van der Meer, and H.-R. Siegrist. 2000. Mathematical modeling of autotrophic denitrification in a nitrifying biofilm of a rotating biological contactor. Water Sci. Technol. 41:191-198. [Google Scholar]
- 21.Koops, H.-P., and A. Pommerening-Röser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 22.Logemann, S., J. Schantl, S. Bijvank, M. van Loosdrecht, J. G. Kuenen, and M. Jetten. 1998. Molecular microbial diversity in a nitrifying reactor system without sludge retention. FEMS Microbiol. Ecol. 27:239-249. [Google Scholar]
- 23.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nogueira, R., L. F. Melo, U. Purkhold, S. Wuertz, and M. Wagner. 2002. Nitrifying and heterotrophic population dynamics in biofilm reactors: Effects of hydraulic retention time and the presence of organic carbon. Water Res. 36:469-481. [DOI] [PubMed] [Google Scholar]
- 25.Pollice, A., V. Tandoi, and C. Lestingi. 2002. Influence of aeration and sludge retention time on ammonium oxidation to nitrite and nitrate. Water Res. 36:2541-2546. [DOI] [PubMed] [Google Scholar]
- 26.Pommerening-Röser, A., G. Rath, and H.-P. Koops. 1996. Phylogenetic diversity within the genus Nitrosomonas. Syst. Appl. Microbiol. 19:344-351. [Google Scholar]
- 27.Princic, A., I. I. Mahne, F. Megusar, E. A. Paul, and J. M. Tiedje. 1998. Effects of pH and oxygen and ammonium concentrations on the community structure of nitrifying bacteria from wastewater. Appl. Environ. Microbiol. 64:3584-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: Implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N. Y.
- 31.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. Jetten, J. W. Metzger, K. H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 32.Schramm, A., D. de Beer, M. Wagner, and R. Amann. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegrist, H. 1996. Nitrogen removal from digester supernatant: comparison of chemical and biological methods. Water Sci. Technol. 34:399-406. [Google Scholar]
- 34.Siegrist, H., S. Reithaar, G. Koch, and P. Lais. 1998. Nitrogen loss in a nitrifying rotating contactor treating ammonium-rich wastewater without organic carbon. Water Sci. Technol. 38:241-248. [Google Scholar]
- 35.Sliekers, A. O., N. Derwort, J. L. Gomez, M. Strous, J. G. Kuenen, and M. S. Jetten. 2002. Completely autotrophic nitrogen removal over nitrite in one single reactor. Water Res. 36:2475-2482. [DOI] [PubMed] [Google Scholar]
- 36.Strous, M., E. van Gerven, J. G. Kuenen, and M. S. M. Jetten. 1997. Effects of aerobic and microaerobic conditions on anaerobic ammonium oxidizing (anammox) sludge. Appl. Environ. Microbiol. 63:2446-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strous, M., J. G. Kuenen, and M. S. Jetten. 1999. Key physiology of anaerobic ammonium oxidation. Appl. Environ. Microbiol. 65:3248-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Third, K. A., A. O. Sliekers, J. G. Kuenen, and M. S. M. Jetten. 2001. The CANON system (completely autotrophic nitrogen-removal over nitrite) under ammonium limitation: Interaction and competition between three groups of bacteria. Syst. Appl. Microbiol. 24:588-596. [DOI] [PubMed] [Google Scholar]
- 39.van Dongen, U., M. S. Jetten, and M. C. van Loosdrecht. 2001. The Sharon-anammox process for treatment of ammonium rich wastewater. Water Sci. Technol. 44:153-160. [PubMed] [Google Scholar]
- 40.van Loosdrecht, M. C. M., and M. S. M. Jetten. 1998. Microbiological conversions in nitrogen removal. Water Sci. Technol. 38:1-7. [Google Scholar]
- 41.Wagner, M., G. Rath, R. Amann, H. P. Koops, and K. H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 42.Wagner, M., G. Rath, H. P. Koops, J. Flood, and R. Amann. 1996. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. 34:237-244. [Google Scholar]