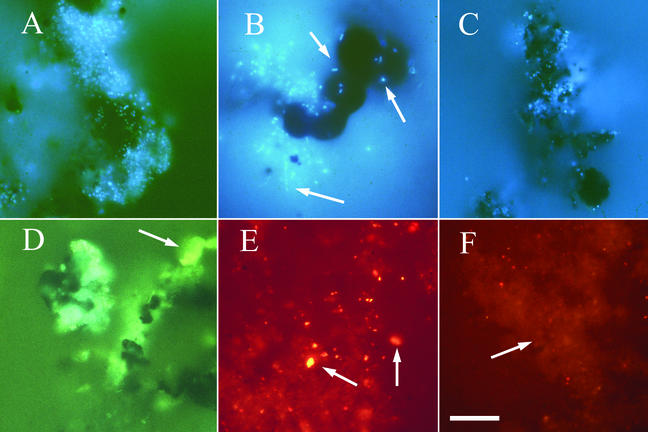
FIG. 3.
Epifluorescence photomicrographs of fluorescently stained microbial communities inhabiting the walls of Finn. (A to C) Images obtained by DAPI staining of attached microbial communities. (D to F) Images obtained by FISH to mineral surfaces and cell extracts. Biofilms were a common feature in Z2 and Z3 (A) and in some cases were more than 10 μm thick. Dispersed cell clusters (B) and microcolonies (C) were also observed within Z2 and Z3, respectively. Attached communities were composed of cells with diverse morphologies, including rods, cocci, and filaments (arrows in panel B). (D) Whole-cell hybridization of a microbial biofilm attached to unextracted rock material from Z3 with ARC915 (green fluorescence). Background fluorescence of the mineral substratum (yellow-green) is indicated by an arrow. (E) FISH to a cell extract from Z2, showing the nonspecific binding of probe ARC915 (red fluorescence) to small mineral particles (indicated by arrows). In this experiment we did not use the blocking agents bovine serum albumin and poly(A), which were typically used in FISH experiments in this study. (F) FISH to a cell extract from Z3 with the crenarchaea-specific probe CREN499 (red fluorescence). The diffuse fluorescence of nonspecific probe binding is indicated by an arrow, in contrast to the point sources of fluorescence (red dots), which were counterstained with DAPI and were counted as hybridized cells. All images were captured with a 35-mm microscope camera, scanned, and processed in Adobe Photoshop, version 6.0. (Adobe). Scale bar = 15 μm.