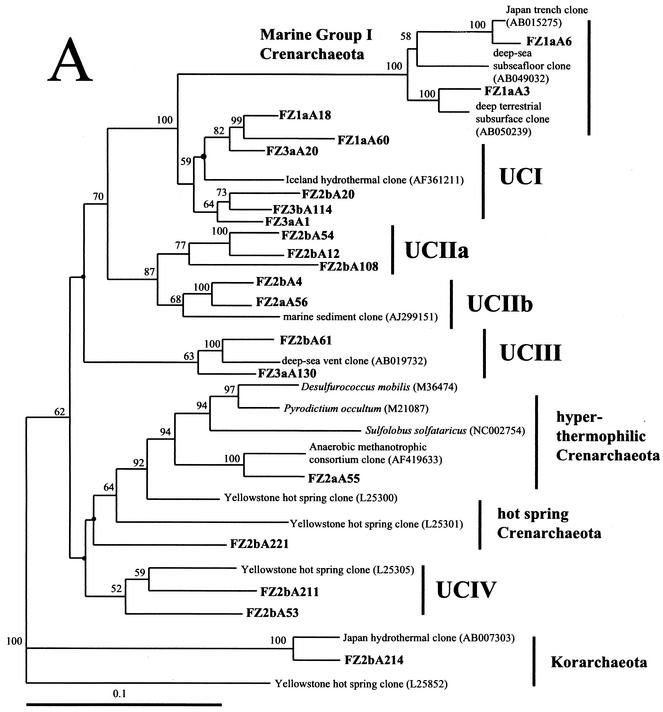
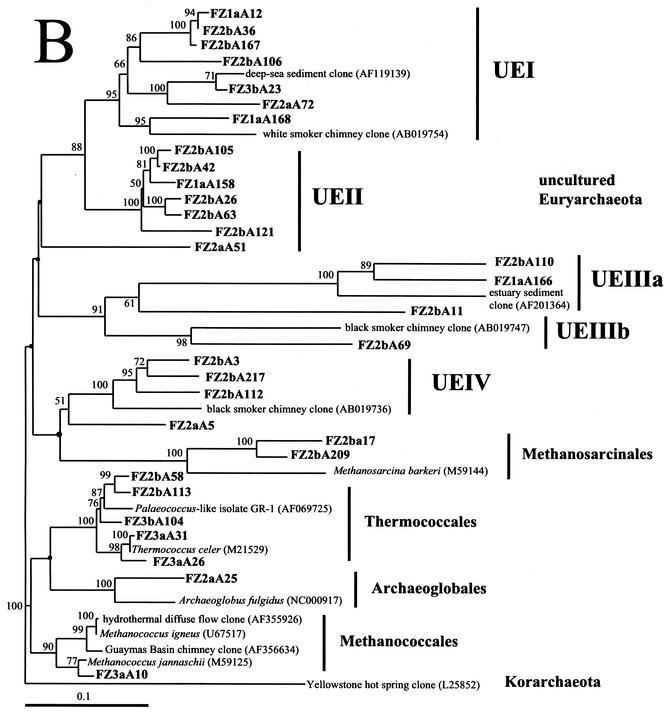
FIG. 5.
Phylogenetic trees based upon 16S rRNA gene sequences of archaea, including clones obtained from Z1 to Z3 of the black smoker chimney Finn. Each of the trees was inferred by phylogenetic analysis of approximately 600 homologous positions of sequences by using the Fitch-Margolis distance-based method. The numbers in parentheses are the GenBank accession numbers for sequences obtained from NCBI. Percentages of 100 bootstrap resamplings greater than 50% are indicated adjacent to the corresponding nodes. The bootstrap values were less than 50% for the branch points marked with solid circles and no numerical values. Scale bars = 0.1 expected nucleotide change per sequence position. (A) Tree depicting the phylogenetic diversity of Crenarchaeota in Z1 to Z3 of Finn. Some clone phylotypes in the marine group I Crenarchaeota were omitted for simplicity. (B) Tree indicating the phylogenetic relationships of Euryarchaeota in Z1 to Z3 of Finn. FZ1a is from Z1 of Finn, FZ2a and FZ2b are from Z2 of Finn, and FZ3a and FZ3b are from Z3 of Finn. Corresponding data are presented in Table 3.