Abstract
Bone marrow transplantation in animals has been shown to generate epithelial populations, a phenomenon that has also recently been suggested to take place after human hematopoietic cell transplantation. However, reports in humans are not conclusive because they still leave open the possibility that the identified donor-derived cells are not epithelial cells but intraepithelial lymphocytes. Here, we demonstrate that donor-derived CD45+ hematopoietic cells in close contact with epithelial tissue may be falsely characterized as donor-derived epithelial cells if the three-dimensional structure of the tissue is not considered and the hematopoietic markers are not examined. By using a rigorous three-dimensional analysis on single sections of colon biopsies triple stained with donor-specific, epithelial-specific, and hematopoietic-specific markers we demonstrate that chimerism of colon epithelium is a real phenomenon occurring constantly after human hematopoietic cell transplantation. We exclude horizontal DNA transfer or cell fusion as the underlying mechanism of our findings. Tissue damage enhances the engraftment of the donor-derived epithelial cells. The physiological and therapeutical role of the donor-derived epithelial cells after human hematopoietic cell transplantation needs further investigation. However, their identification requires stringent and unequivocal detection systems.
Hematopoietic cell transplantation (HCT) is a standard term used in clinical practice indicating a procedure in which a cellular graft usually derived from bone marrow (BM) or peripheral blood stem cells reconstitutes the hematopoietic system in a myeloablated host. Allogeneic HCT results in true biological chimeras. Although circulating hematopoietic cells and their tissue derivatives such as Kupffer cells in the liver,1 Langerhans cells in the skin,2 and microglial cells in the brain3 become donor genotype after transplantation, other cells are believed to remain recipient in origin. However, studies in laboratory animals throughout the last few years indicate that bone marrow transplantation (BMT) generates unexpected populations in vivo, such as liver cells and other epithelial cells.4–8 More recently, genetic marker studies in humans suggested generation after HCT of donor-derived hepatocytes, skin cells, and epithelial cells of the gastrointestinal tract.9–12 However, reports in humans are not conclusive because of methodological limitations that still leave open the possibility that the identified donor-derived cells are not epithelial cells but intraepithelial lymphocytes, and therefore have been treated with skepticism.13–15
We sought to find out if human HCT results in generation of donor-derived epithelial cells. Attention has been paid to the technical aspects to be sure that the identified donor-derived cells express epithelial cell markers and are not contaminating hematopoietic cells. Frozen colonic biopsies of eight female patients who underwent a sex-mismatched allogeneic HCT were triple-stained by combining interphase fluorescence in situ hybridization (FISH) with a Y-chromosome-specific probe, immunofluorescent labeling for cytokeratins (CKs) and either TOTO-3 nuclear label or CD45 immunofluorescence. The samples were examined by confocal microscopy and then underwent a rigorous three-dimensional analysis. Here, we demonstrate that donor-derived CD45+ lymphocytes in close contact with epithelial tissue may be falsely characterized as donor-derived epithelial cells if the three-dimensional structure of the tissue is not considered and the hematopoietic markers are not examined. We found that chimerism of colon epithelium is a real phenomenon occurring constantly after human HCT. Three-dimensional X-chromosome enumeration in individual Y+ epithelial cells was performed to examine whether or not fusion is the underlying mechanism of the chimeric events found.
Materials and Methods
Patients and Tissue Specimens
Clinical information regarding the HCT recipients, their diagnosis, the history of male childbearing, the type of transplantation (BMT or peripheral blood stem cell transplantation) and the time from transplantation to colon tissue sampling is summarized in Table 1. BM recipients received between 190 to 280 × 108 mononuclear cells and peripheral blood stem cell recipients between 743 to 1010 × 108 mononuclear cells. In patients showing clinical signs of gastrointestinal graft versus host disease at different time points after transplantation, endoscopic biopsies were taken after informed consent to verify the diagnosis. Snap-frozen biopsies obtained from eight female patients who underwent a sex-mismatched allogeneic HSCT as well as from six female and two male sex-matched transplanted patients were stored at −180°C after informed consent. Other biopsies obtained from the same endoscopy were formalin-fixed and examined in the Pathology Department of the University of Freiburg. Cryostat sections used in this study were examined after informed consent. All patients had received multiple blood transfusions in the past; blood products transfused after transplantation were always leukocyte depleted and irradiated.
Table 1.
Patient Characteristics and Colon Epithelium Chimerism
| Pt | Recipient/ Donor | Male child-bearing | Age at HCT (years) | Reason for HSCT | Type of HSCT | Days from HSCT to biopsy | Pathological diagnosis | Intraepithelial chimeric events/ 100 crypts
|
% of intraepithelial Y+/total epithelial cells | % of Y+/CK+/ CD45−/total epithelial cells | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y+/CD45+ | Y+/CK+/CD45− | Total | ||||||||||
| Sex-mismatched | ||||||||||||
| 1 | F/M | No | 41 | CML | BM+PBSC | 420 | GvHD I | 39 | 12 | 51 | 0.80% | 0.19% |
| 2 | F/M | Yes | 43 | CML | BM | 29 | Intact epithelium | 1 | 3 | 4 | 0.06% | 0.05% |
| 3 | F/M | Yes | 29 | CML | BM | 43 | GvHD III | 30 | 11 | 41 | 0.65% | 0.17% |
| 4 | F/M | No | 28 | AML | BM | 72 | GvHD I | 22 | 24 | 46 | 0.73% | 0.38% |
| 5 | F/M | Yes | 51 | CML | PBSC | 16 | Intact epithelium | 1 | 2 | 3 | 0.05% | 0.03% |
| 6 | F/M | Yes | 38 | AML | BM/PBSC* | 540† | GvHD III | 33 | 19 | 52 | 0.82% | 0.30% |
| 7 | F/M | No | 22 | CML | BM | 41 | GvHD IV | 17 | 14 | 31 | 0.49% | 0.22% |
| 8 | F/M | No | 48 | AML | PBSC | 15 | GvHD I | 9 | 6 | 15 | 0.24% | 0.09% |
| Sex-matched | ||||||||||||
| 9 | F/F | No | 19 | ALL | PBSC | 19 | GvHD I | Y− | NA | |||
| 10 | F/F | Yes | 62 | AML | BM | 40 | GvHD II | Y− | NA | |||
| 11 | F/F | Yes | 34 | Mel | PBSC | 20 | GvHD III | Y− | NA | |||
| 12 | F/F | Yes | 42 | Ly | PBSC | 231 | GvHD III | Y− | NA | |||
| 13 | F/F | Yes | 44 | CML | PBSC | 60 | Intact epithelium | Y− | NA | |||
| 14 | F/F | Yes | 44 | ALL | PBSC | 20 | GvHD I | Y− | NA‡ | |||
| 15 | F/F | No | 67 | AML | PBSC | 51 | Intact epithelium | Y− | NA‡ | |||
| 16 | M/M | - | 18 | AML | PBSC | 30 | Intact epithelium | 3 | >99% | NA | ||
| 17 | M/M | - | 55 | Ly | PBSC | 45 | GvHD III | 23 | >99% | NA | ||
Eight-μm sections were used for quantification.
BMT followed by PBSCT 5 months later.
Time after the BMT.
Y-FISH was performed in paraffin-embedded materials.
Y−/+, Y-chromosome negative/positive; NA, epithelial chimerism analysis not applicable; BM, bone marrow; PBSC, peripheral blood stem cells; AML/CML, acute-/chronic-myeloid leukemia; Mel, melanoma; Ly, lymphoma; GvHD, graft versus host disease.
Triple Stainings: Combination of FISH for Y- or XY-Chromosomes, Immunofluorescence Staining for CK, and TOTO-3 Nuclear or CD45 Immunofluorescence Staining
Cryostat sections (8 μm except otherwise indicated) were fixed in cold methanol/acetone solution (1:1) for 10 minutes, and air-dried. The sections were pretreated with ribonuclease A (RNase A) (2 mg/ml at 37°C for 45 minutes; Roche Diagnostics, Indianapolis, IN USA) and then processed at 37°C for 30 minutes in preheated 2× standard saline citrate (SSC)/0.1% Nonidet P-40 buffer (Calbiochem, La Jolla, CA), pH 7.0. Serial ethanol dehydration was done (70%, 85%, and 100%; 1.5 minutes each), and the slides were air-dried at room temperature. One μl of the human Y-chromosome-specific probe labeled with rhodamine (CEP Y satellite III Spectrum Orange; Vysis, Bergisch-Gladbach, Germany) was mixed with 7 μl of hybridization buffer (CEP hybridization buffer containing dextran sulfate, formamide, and SSC; Vysis) and 2 μl of purified H2O and applied to the sections. In the case of the combined XY-chromosome stain, a XY mixed probe was used (Vysis). The sections were coverslipped and sealed with rubber cement. The sections and the probe were co-denaturated by placing the slides on the surface of a 73°C prewarmed plate (HYBrite, Vysis) for 3 minutes. Hybridization was performed overnight at 37°C in a humidified box. The next day, the coverslips were carefully removed in 2× SSC/0.3% Nonidet P-40 buffer at room temperature and posthybridization washes were done three times for 2 minutes each in 2× SSC/0.3% Nonidet P-40 preheated buffer at 73°C. For combined Y-chromosome FISH and CK fluorescence immunohistochemistry, sections were then incubated for 30 minutes at room temperature in 1% normal bovine serum (30% albumin bovine solution; Sigma, Germany) diluted in phosphate-buffered saline (BioWhittaker, Germany) to block nonspecific binding. After washing in 2× SSC, the sections were exposed for 3 hours at 37°C to a fluorescein isothiocyanate (FITC)-labeled, monoclonal mouse antibody that detects human CK7 and CK8 (clone CAM 5.2; Becton Dickinson, San Jose, CA). For those sections without CD45 staining, the sections were stained with the nucleic acid dye TOTO-3 as a nuclear counterstain. After a thorough wash in SSC/0.1% Tween 20, the sections were treated with ribonuclease A (see above) and processed with TOTO-3 nucleic acid dye (4 × 10−4 mmol/L, at room temperature, for 10 minutes; Molecular Probes, Leiden, The Netherlands). For the combination of FISH for Y-chromosome and immunofluorescence for CD45 and CK, after posthybridization washes and antigen blocking, sections were first incubated with a CD45 primary antibody (monoclonal mouse anti-human LCA, clones 2B11+ PD7/26, 16 μg/ml, at room temperature for 1 hour; DAKO, Glostrup, Denmark), washed in 2× SSC/0.1% Tween and then exposed to a goat anti-mouse antibody labeled with Alexa Fluor 633 [Alexa Fluor 633 F(ab′)2-fragment of goat anti-mouse IgG (H+L), 10 μg/ml, at room temperature for 1 hour; Molecular Probes]. These steps were followed by the CK staining procedure as described above. After washing in SSC/0.1% Tween 20, slides were mounted with Vecta-shield anti-fade (Vector Laboratories, Burlingame, CA) and the coverslip was added. Negative controls included omission of the Y-probe in FISH, use of isotype-matched control antibodies (IgG1-FITC; Immunotech, Marseille, France) for CK and CD45 staining, and staining of gut samples from female patients who received BM transplants from a female donor. Positive controls consisted of samples obtained from transplanted male BM recipients.
The sensitivity of our Y/CK/TOTO or Y/CK/CD45 stain to identify Y signals was tested in control male biopsies and was found to be dependent on the thickness of the section (see Results). By using 8-μm sections the sensitivity was found to be >98% for the Y-signal. Nearly all of the TOTO-stained nuclei in the 20-μm male sections were found to be Y-positive indicating that in these thicker sections our FISH technique did not lose hybridization efficiency. Combined XY FISH analysis (X/Y/CK stain) was tested in 20-μm control male and female sections. Despite the thickness of these sections, expression of the signals was consistent throughout the whole tissue. Individual cells from female and male controls were tested with three-dimensional analysis and found to contain either XX or XY signals, respectively.
Immunohistochemistry for CD45 (LCA) and CD68
The adjacent serial sections (8 μm or 4 μm) below and above the section that was used for FISH Y-chromosome analysis (middle section) were fixed with acetone and stained with CD45 (LCA)-specific antibodies (monoclonal antibody, clone 2B11+PD7/26; DAKO-Diagnostica, Hamburg, Germany). Antibody binding was detected at room temperature using affinity-purified rabbit anti-mouse IgG (1:25) for 30 minutes and mouse alkaline phosphatase anti-alkaline phosphatase complex (1:50) for 30 minutes (DAKO). Two percent normal pooled human serum was added to rabbit anti-mouse IgG. Incubations with the secondary antibodies and the alkaline phosphatase anti-alkaline phosphatase complex were repeated once. Bound alkaline phosphatase was demonstrated using naphthol AS-Bi phosphate (Sigma, St. Louis, MO) as a substrate and New Fuchsine (Serva, Heidelberg, Germany) as a coupling reagent in 0.2 mol/L of Tris-HCl buffer, pH 8.5) with 1 mmol/L of levamisole in the reaction mixture to block endogenous enzyme activity. The slides were counterstained with hematoxylin and mounted. Those colonic crypts that were found to contain Y+/CK+ cells in the triple-stained middle section were identified in the CD45-labeled sections and color photographed at various magnifications (×10, ×63, and ×100). Serial frozen sections were stained with CD68-specific antibodies (monoclonal antibody, clone PG-M1; DAKO-Diagnostica) by using the alkaline phosphatase anti-alkaline phosphatase technique as described above.
Tissue and Image Analysis
Confocal laser-scanning microscopy was performed using a Zeiss LSM 410 scanning confocal microscope (Zeiss, Jena, Germany). The Y-chromosome rhodamine signal was excited at 543 nm and emission was collected from 590 to 610 nm whereas the CK FITC signal was excited at 488 nm and emission collected from 510 to 525 nm. TOTO-3 and Alexa 633 were excited at 633 nm while emission was collected above 665 nm. For clarity, in those samples in which CD45-Alexa 633 was present, the CK-FITC signal was shown on the blue channel of the microscope. This allowed the signal of the CD45-Alexa 633 to be more easily seen on the relatively brighter green channel. Between 60 to 100 serial confocal images of suspected cells were collected at an interval of from 0.12 to 0.2 μm along the z-axis with the 1.2 numerical aperture ×40 water immersion or the 0.5 numerical aperture ×20 objective. The serial two-dimensional images collected through the thickness of the specimen were then processed to construct three-dimensional projections with the use of a specialized commercial software (Autovisualize 5.5; AutoQuant Imaging). To minimize the possible influence of spherical aberration of the signals on our analysis, three-dimensional co-localization of the Y-chromosome and CK was considered positive only if at least two-thirds of the Y-chromosome signal was surrounded by the CK label.
Quantitative Data of Epithelial Cell Engraftment
At least five frozen sections from each patient were examined with the Y-chromosome/CK/TOTO-3 or the Y-chromosome/CK/CD45 triple stain. To obtain quantitative data of epithelial cell engraftment, at least 100 large bowel crypts triple stained with Y/CK/CD45 were examined for each patient. In the more than 500 crypts that were carefully examined, we found that every crypt contained between 39 to 73 epitheliocytes (mean, 63 epitheliocytes), regardless of the presence of GvHD. Therefore, examination of 100 crypts results in an evaluation of a mean 6300 epitheliocytes (range, 3900 to 7300 epitheliocytes) in every patient. The degree of epithelial chimerism was presented as the absolute number of Y+/CK+/CD45− cells/100 bowel crypts examined or as the calculated mean percentage of Y+/CK+/CD45− cells/total epitheliocytes by using the formula: Y+/CK+/CD45− cells/total epithelial cells in % = (absolute number of Y+/CK+/CD45− cells/100 bowel crypts × 100)/6300.
Results
Characteristics of Patients: Specificity and Sensitivity of the FISH Y-Chromosome Analysis
Clinical information regarding the HCT recipients, their diagnosis, the history of male childbearing, the type of transplantation (BMT or peripheral blood stem cell transplantation), and the time from transplantation to colon tissue sampling is summarized in Table 1. Fresh frozen colon biopsies were in situ hybridized with a human Y-chromosome-specific probe. Male and female patients who had received sex-matched HCT were used as controls. In these samples Y-chromosome was only detected in the male intestine, demonstrating hybridization specificity (Figure 1, Table 1). We never detected any Y-chromosome in biopsies taken from seven sex-matched transplanted women used as controls, despite the fact that five of them had a history of male childbearing and all of them received multiple blood transfusions in the past (Table 1). The FISH Y-signal was localized within the TOTO-3-counterstained nuclei representing true Y-chromosomal material rather than nonspecific debris.
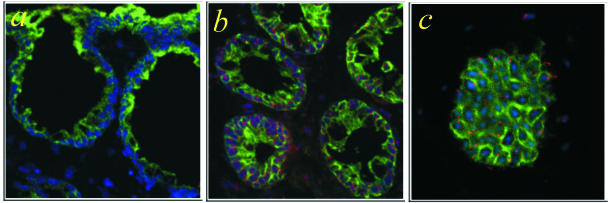
Figure 1.
Specificity and sensitivity of the FISH Y analysis. Triple staining of colon biopsies taken from HCT recipients by using FISH for the visualization of the Y-chromosome (red), fluorescence immunohistochemistry for CK (CK, green), and TOTO-3 nuclear counterstain (blue). Two-dimensional images of triple-stained colon biopsies from a female recipient after HCT from a female donor (a) and a male recipient after HCT from a male donor (b, c). In a and b 8-μm sections were stained, in c 4-μm sections were stained. The Y-chromosome was restricted to the male intestine, demonstrating hybridization specificity. Because of the partial sampling of the cells in thinner sections, the sensitivity of the FISH Y analysis was >98% in the 8-μm sections and ∼75% in the 4-μm sections.
Epithelial cells have a diameter of ∼10 to 15 μm. Therefore, thin sections may result in a partial sampling of the cells. We tested the sensitivity of our FISH technique to identify Y-chromosome cells on sections of different thicknesses. In the control 4-μm male thin sections we found the Y-signal in ∼75% of the cells (Figure 1c). However, by using 8-μm sections we could reliably find Y-signal in >98% of the cells (Figure 1b). Therefore, 8-μm sections were used in this study.
Donor-Derived Epithelial Cell or Donor-Derived Cell Lying Above an Epithelial Cell?
To detect donor-derived epithelial cells in female sex-mismatched HCT recipients we performed triple staining by combining FISH-Y labeling, immunofluorescent labeling for CKs, and TOTO-3 nuclear counterstain within a single 8-μm colon section. By examining the FISH-Y/CK/TOTO-stained sections with two-dimensional microscopy we identified Y-chromosome-positive (Y+) cells that appeared to be also CK+ in all eight patients examined (Figure 2). However, by using confocal microscopy and a three-dimensional analytical method we found that two-dimensional microscopy may misrepresent critical information about the three-dimensional distribution of the examined markers. Overlapping cells produced artifacts showing in the two-dimensional analysis what appears to be marker co-localization within a single cell, although the markers are actually expressed in different cells. This phenomenon is clearly illustrated in Figure 2, a and b. In the two-dimensional image of a colonic crypt some CK+ cells seem to display the Y-chromosome (Figure 2, a and b; left). However, the collection of serial optical sections through the thickness of the specimen by laser-scanning confocal microscopy and their three-dimensional reconstruction demonstrates a missing co-localization of the CK and Y-chromosome signals in the z-dimension (Figure 2, a and b, right; and Movie 2a, which is published as supporting information on The American Journal of Pathology web site). The Y-chromosome-specific signals could belong to a donor-derived lymphocyte lying above or below the CK+ cell, leading to a misinterpretation of the two-dimensional micrographs. Of a total of 63 chimeric events that were detected with two-dimensional microscopy as intraepithelial, we found that on examination in three-dimensional microscopy 19 of these cases were not truly intraepithelial. Thus, compared to three-dimensional microscopy two-dimensional microscopy of 8-μm sections results in ∼30% false-positive events.
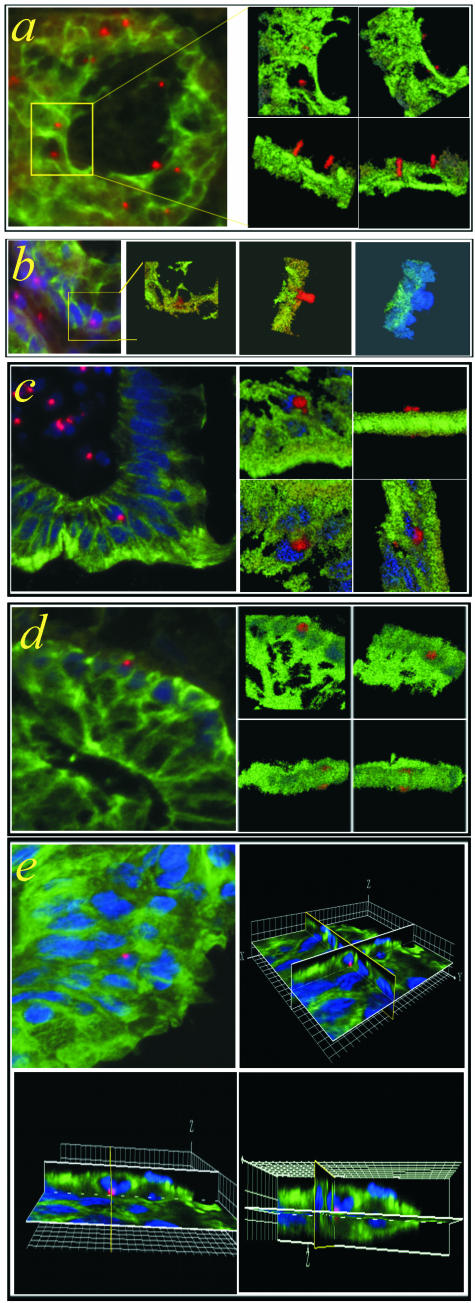
Figure 2.
Pitfalls by the detection of epithelial chimerism: overlapping cells? FISH Y-chromosome (red)/CK (green)/TOTO-3 (blue) triple-immunofluorescence staining of colon biopsies (8-μm sections) from female recipients transplanted with male BM cells. Two-dimensional images and three-dimensional volume projections at different angles are shown. a and b: The two-dimensional micrographs (left) of intestinal villi suggest the presence of CK+ cells containing the Y-chromosome (boxed areas). The collection of serial optical sections through the thickness of the specimen with the use of scanning laser confocal microscopy and their three-dimensional reconstruction demonstrates, however, a missing co-localization of the CK and the Y-chromosome signals in the z-dimension (right). In b the cells are shown in separate panels both with and without the TOTO-3 nuclear stain (blue). c and d: Three-dimensional co-localization of CK and Y-chromosome, strong CK immunostaining, and presence of the Y-chromosome signal within the TOTO-3-labeled nucleus. The absence of green CK staining at the top or the bottom of the three-dimensional projections shown in c and d is because of the partial sampling of the cells in the 8-μm tissue sections. e: Two-dimensional (top left) and three-dimensional views of a 20-μm tissue section. Compared to the thinner 8-μm sections, in this 20-μm-thick section entire cells are contained as can be seen in the Z-plane of the three-dimensional reconstruction. Because the CK skeleton completely surrounds the internal structures, the intracellular signals can be difficult to see with three-dimensional volume projections. Therefore, to more clearly demonstrate the signals within the cell, the three-dimensional reconstruction shown in e uses orthogonal slices in all three dimensions, centered on the Y-chromosome signal. The Y-chromosome is clearly confined to the blue nucleus, which itself is fully contained within the green CK structure. The three-dimensional images show the same cell as it appears from different angles of view. Donor-derived epithelial cells or intraepithelial lymphocytes enclosed by epithelial cells? Three-dimensional movie projections are published as supporting online material on The American Journal of Pathology web site (movies 2; a, c, d, e).
The absence of green CK staining in the top or the bottom of the three-dimensional projections shown in Figure 2, c and d, is because of the partial sampling of the cell in the 8-μm tissue sections. In contrast, in Figure 2e the Y-chromosome-positive nucleus is surrounded by a CK-positive cytoskeleton as a result of the sampling of an entire epithelial cell located within the 20-μm tissue section. Thick 20-μm sections were examined from a total of five patients and completely sampled CK+/Y+/TOTO-3 cells were found in each case.
Donor-Derived Epithelial Cell or Intraepithelial Lymphocyte?
By using the FISH-Y/CK/TOTO triple stain and the three-dimensional analysis we found cells that appeared to be donor-derived Y+/CK+ epithelial cells in all eight patients examined (Figure 2; c, d, and e). However, with this stain we could not exclude the possibility that the intraepithelial found Y-chromosome signal was from infiltrating lymphocytes enclosed within the epithelial tissue. We established a triple fluorescence stain by combining FISH Y-chromosome labeling (red) with CK (blue) and CD45 (green) immunofluorescence. Donor-derived lymphocytes stained positive as expected for both the Y-chromosome and CD45. We found that intraepithelial lymphocytes can be easily mistaken as donor-derived epithelial cells if the expression of hematopoietic markers is not examined, as shown in Figure 3a.
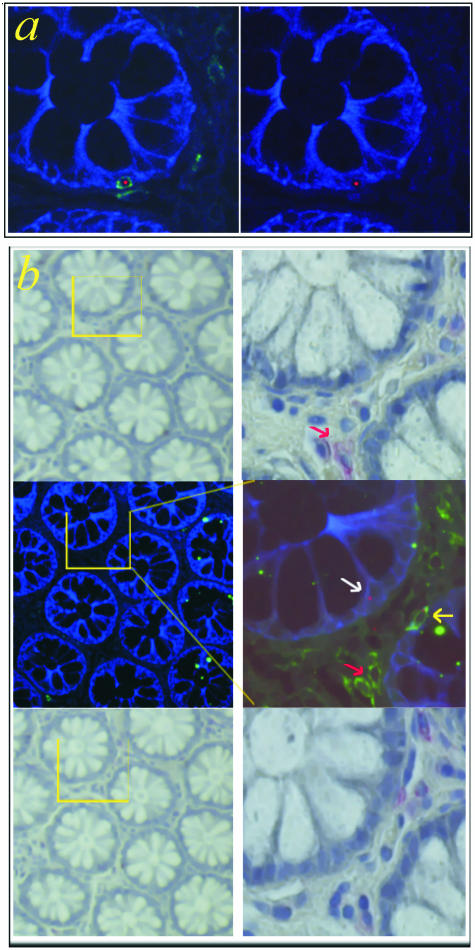
Figure 3.
Pitfalls by the detection of epithelial chimerism: donor-derived epithelial cell or intraepithelial lymphocyte? a: Triple staining of colon biopsies taken from HCT female recipients by using FISH for the Y-chromosome (red) and fluorescence immunohistochemistry for CK (blue) and for CD45 (green). A donor-derived intraepithelial lymphocyte detected with this method may falsely be detected as Y+/CK+ cell if CD45 expression is not examined (right, green channel turned off). b: Staining of CD45 in serial 4-μm sections cannot exclude the presence of lymphocytes in the section studied by FISH analysis. The middle 4-μm section (middle row) was stained with the Y (red)/CK (blue)/CD45 (green) triple stain, while the adjacent sections (4 μm) above (top row) and below (bottom row) are immunocytochemically stained with CD45 antibodies (red). The intraepithelial CD45+ cell (yellow arrow) visible in the Y/CK/CD45-stained middle section is not detected in the serial sections above or below. Other CD45+ cells located in the lamina propria that were detected in the middle and an adjacent section are not apparent in the remaining section (red arrow). A Y+/CK+/CD45− donor-derived epithelial cell is also seen (white arrow). Left and right, low- and high-power magnification, respectively.
Staining of CD45 in serial 4-μm sections, as has been used in studies to date, cannot exclude the presence of lymphocytes in the section studied by FISH Y analysis. As is clearly illustrated in Figure 3b, CD45+ cells found in one section (4 μm thick) are not necessarily detectable in the neighboring sections (4 μm each) below and above. For example, the intraepithelial lymphocyte found in the Y/CK/CD45-stained middle section is not detected in the serial sections above or below. In the same vein, other CD45+ cells that were stained in the middle and the one adjacent section are not apparent in the other section.
Donor-Derived Epithelial Cells in Sex-Mismatched HCT Recipients
We used the triple FISH/CK/CD45 stain in a single 8-μm section combined with three-dimensional analysis to detect donor-derived epithelial colon cells in the sex-mismatched HCT recipients. As expected, the majority of the intraepithelial chimeric events found were because of Y+/CD45+ intraepithelial lymphocytes (Table 1). However, we could also detect Y+/CK+/CD45− cells in all eight patients (Figure 4 and Figure 3b). In all cases examined, Y+/CK+/CD45− cells were identified as isolated single cells scattered throughout the colonic crypts and not as clusters. At least 100 large bowel crypts were examined for each patient. Up to 24% of the crypts were found to contain single epithelial cells of donor origin (patient 4). Considering that each crypt contains a mean of 63 epitheliocytes, we conclude that a mean of 0.18% of the overall colon epithelial cells were donor-derived, with higher numbers found in the sections with histologically documented tissue damage (mean, 0.22%; six patients) compared to sections with intact epithelium (mean, 0.04%; two patients). Tissue macrophages may down-regulate their CD45-expression and therefore escape their detection as hematopoietic cells. Although CD68-positive macrophages could be identified in the lamina propria, we never detected macrophages within the colon epithelium (data not shown).
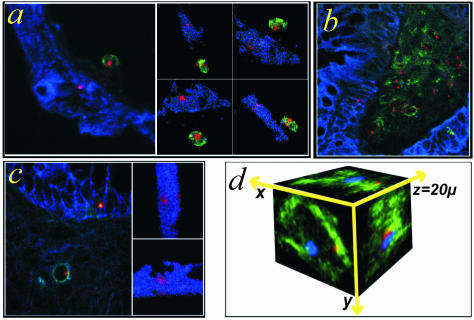
Figure 4.
Epithelial chimerism after human HCT. a–d: FISH Y-chromosome (red)/CK (blue)/CD45 (green) triple-immunofluorescence staining of female colon biopsies (8-μm sections). Two-dimensional and three-dimensional images of Y+/CK+/CD45− cells from different patients. Donor-derived lymphocytes were stained Y+/CD45+, as expected. Three-dimensional movie projections are published as supporting online material on The American Journal of Pathology web site (movies 4, a and c). e: FISH Y-chromosome (blue)/FISH X-chromosome (red)/CK (green) triple-immunofluorescence staining. Images show maximal projections of a CK+/Y+ cell found in a female colon biopsy in all dimensions. A CK+ cell visible in the XY plane contains a Y-chromosome (blue) and only one X-chromosome (red). These chromosomes were also the only ones detected within the cell in the other dimensions as shown.
Donor-Derived Epithelial Cells or Fusion Events?
We examined whether or not the chimeric events found within the gut epithelium are the products of cell fusion. We combined CK staining with FISH for the Y- and the X-chromosome and examined the number of X-chromosomes present within the CK+/Y+ cells. To be sure that we did not underestimate the number of X-chromosomes within a single cell, we used 20-μm sections for these stains to sample whole epithelial cells (diameters, 10 to 15 μm) with their entire nucleus and chromosome content. For correct enumeration of the X signals within a single cell we examined the CK+/Y+ cells in all three dimensions. This was done because examination by confocal microscopy in only a single plane may result in an underestimation of the FISH signals. Individual Y+/CK+ epithelial cells from three female recipients were found to contain only one X-chromosome (Figure 4b). Furthermore, we examined the TOTO-3 nuclei signal in the Y+/CK+/TOTO-3 triple-stained cells in all patients, but we never found any doubled TOTO-3 signal intensity indicative for fusion events.
Discussion
BMT in animals has been shown to generate unexpected populations in vivo, such as liver cells and other epithelial cells.4–8 More recently, Y-chromosome marker studies have suggested that this phenomenon is also taking place after human HCT.9–12 However, reports in humans are still inconclusive because of methodological limitations. The possibility that the identified donor-derived Y+ cells are not epithelial cells but rather donor-derived hematopoietic cells in close contact with epithelial tissue could not be excluded in the human studies reported up to now. The fact that tissue is a compact, three-dimensional structure has not been considered in the interpretation of results from microscopic examinations.16,17 By using confocal microscopy and a three-dimensional analytical method in the present work we clearly demonstrated that donor-derived cells in close contact with epithelial tissue may be falsely characterized as donor-derived epithelial cells when two-dimensional microscopy is used as the method of tissue examination. Furthermore, we demonstrated that infiltrating lymphocytes enclosed within epithelial tissue can be mistaken as donor-derived epithelial cells if the expression of hematopoietic markers is not examined. To exclude lymphocytes as the source of the Y-chromosome signal found within the FISH Y-stained area, Körbling and colleagues11 and Okamoto and colleagues12 stained a 4-μm or 6-μm serial section with CD45 antibodies. However, we clearly demonstrated that examination of serial 4-μm sections cannot exclude the presence of lymphocytes in the Y-chromosome-positive area. Therefore, all of the data reported up to now suggesting generation after human HCT of donor-derived hepatocytes,9–12 skin cells, and epithelial cells of the gastrointestinal tract11,12 by using two-dimensional microscopy and/or examination of adjacent sections are not conclusive, and have been treated with skepticism.13–15
We studied colonic biopsies from eight female patients that underwent an allogeneic HCT from a male donor for the presence of epithelial cells of donor origin. In our study attention has been paid to the technical aspects to be sure that the identified Y+ male cells express epithelial cell markers and do not express hematopoietic markers. By using a rigorous analytical method combining a triple stain in a single section with three-dimensional analysis we unequivocally demonstrate that after HCT in humans, donor-derived cells yield cells that enter the gut and express epithelial-specific features. Both CD45+ and CD68+ cells were excluded as the source of the intraepithelially located Y-chromosome. Because CD45 is a pan-hematopoietic marker, it seems unlikely that the Y-chromosome belongs to an inflammatory cell enclosed within the colon epithelium. We found Y+/CK+/CD45− cells in all eight HSC-transplanted women. Although up to 24% of the intestinal crypts were found to contain donor-derived epithelial cells, only 0.18% of the overall colon epithelial cells was donor derived. On the other hand, partial sampling of the nuclei in the 8-μm sections may have resulted in an underestimation of the degree of epithelial cell engraftment and 20-μm sections would have been more accurate for quantitation purposes. However, because less than 2% of Y+ nuclei are missed when examining 8-μm control male samples, we believe that the use of 8-μm sections in this study has not dramatically influenced the measured degree of epithelial chimerism. Previous studies in humans suggested an incidence of donor-derived epithelial cells up to 17% in the liver10 and up to 3.6% in the gastrointestinal tract.11 Probably the higher incidence of epithelial chimerism found in these studies is because of enumeration not only of the donor-derived epithelial cells but also of the intraepithelial lymphocytes.
Tracking of donor-derived cells in our study was done by detecting the Y-chromosome by FISH. Other possible mechanisms that could explain the presence of Y-chromosome material in female tissue are horizontal DNA transfer through fetal-maternal circulation18 and/or blood transfusion,19 or fusion between donor-derived cells with recipient epithelial cells.20,21 However, we never detected any Y-chromosome in biopsies taken from seven women used as controls, although five of them had a history of male childbearing and all of them received multiple blood transfusions in the past. In addition, four of the women with Y+ cells in their colon epithelium were either nulliparous (three patients) or had only female children (one patient). Thus, Y+ epithelial cells found in the female recipients of transplanted male cells are most likely to be donor in origin. Individual Y+ epithelial cells from three female recipients were found to contain only one X-chromosome. This indicates that at least some of the identified Y+ epithelial cells in the eight women of our study represent donor-derived epithelial cells rather than fusion between donor cells and host epithelial cells. However, fusion events cannot be ruled out because chromosomes could be lost after cell fusion. Cell fusion has been shown to be the principal source of BM-derived hepatocytes in regenerating livers in mice.22,23 In man, nonfusion mechanisms have been implicated in the generation of BM-derived buccal epithelial cells.24
We found donor-derived epithelial cells in all eight patients studied. It may be speculated that the dysfunction of the intestinal tract, which was what originally prompted us to take the colon biopsy, had an influence on the establishment of the epithelial microchimerism. However, no tissue damage was found histologically in two of eight of our patients. To date, the published scientific literature indicates that epithelial microchimerism after animal or human HCT occurs in all transplanted recipients and in all tissues examined, irrespective of the presence of tissue damage.4–12 Thus, it seems that epithelial chimerism after HCT could be a constant event. Because we could not analyze biopsies from the same patient at different times after transplantation we cannot say if the establishment of epithelial microchimerism is a transient phenomenon. This latter possibility is unlikely because we found donor-derived epithelial cells as early as 16 days after transplantation and as late as 540 days after transplant. Because all biopsies were taken after the patients showed hematopoietic regeneration it is not clear whether the epithelial microchimerism was caused by lodging of donor-derived cells in the gut at the time of intravenous injection or by endogenous seeding of cells from the engrafted BM. The experiments described here do not determine which cell(s) within the graft actually gives rise to epithelial cells. Cellular infusions used in clinical practice are heterogeneous and contain various types of cells. The cells in question could be as-yet-unknown nonhematopoietic progenitors, mesenchymal progenitors, or hematopoietic or endothelial progenitors with the ability to switch fates.
Y+/CK+/CD45− cells were found in colon epithelium only focally as isolated single cells scattered throughout the colonic crypts. If a donor-derived cell would generate an intestinal epithelial stem cell one would expect large clones of cells within individual villi to carry the Y-chromosome. The pattern we found rather suggests that a donor-derived cell directly forms an epithelial cell. This observation would be consistent with the recently proposed “Chiaroscuro” stem cell model.25 Rather than a hierarchical structure of the hematopoietic (or other) system, this model suggests a very flexible system in which phenotypic shifts, probably also from hematopoietic to epithelial, could sequentially occur dependent on cell-cycle phase and specific microenvironment.
Whether epithelial chimerism after human HCT is an incidental by-product of transplantation without ancillary biological significance or has clinical consequences is still unknown. Because this phenomenon occurs only focally it may be speculated that it does not result in any clinical effects. However, parenchymal microchimerism has been implicated in the pathogenesis of autoimmune diseases26–28 or vascular rejection of human kidney allografts.29 Interestingly, we found more engrafted crypts and more donor-derived epithelial cells in the biopsies showing tissue damage (mean 0.22%) compared to the biopsies with intact epithelium (mean, 0.04%). Despite the small patient numbers, this finding indicates that during tissue damage the repopulation rate of the epithelium with donor-derived epithelial cells is increased. Whether this repopulation has any biological implications and which mechanisms are behind it remains unclear. Because we did not find any clusters of Y+ epithelial cells and the degree of engraftment in a single crypt appears to be fairly constant it seems unlikely that, at least in the setting of the clinical HCT, the donor-derived cells functionally contribute to tissue repair by replacing damaged epithelia. During the review process for this study, an elegant study appeared suggesting that HCT in mice results in epithelial chimerism in damaged pancreas and that HCT contribute to tissue repair by initiation of the endogenous regeneration process.30 Further studies in humans are needed to explore if this is also true in humans and if donor-derived epithelial cells are involved in this. The transcription profiles of the donor-derived epithelial cells after HCT and the characterization of the special microenvironments in which they integrate is likely to answer these questions and may elucidate regulatory cell fate mechanisms providing novel tools for reparative cell or gene therapies. However, as we have demonstrated here, identification of donor-derived epithelial cells for their further characterization is not straightforward and requires stringent and unequivocal detection systems such as the use of combined stains within a single section and the use of laser-scanning confocal or deconvolution microscopic methods.
Supplementary Material
Acknowledgments
We thank Mrs. L. De Lima-Hahn and Mrs. B. Weinhold for technical assistance; Dr. F. Rosenthal and Dr. C. Waller for helpful discussions; and Prof. R. Mertelsmann for supporting this study, critical discussions, and contributions to the manuscript.
Footnotes
Address reprint requests to Dr. Alexandros Spyridonidis, Freiburg University Medical Center, Hugstetterstrasse 55, 79106 Freiburg, Germany. E-mail: spyridonidis@mm11.ukl.uni-freiburg.de.
Supported by a grant from the Landesstiftung Baden Württemberg (to A.S.).
Supplemental material is available at http://www.amjpathol.org.
References
- Gale RP, Sparkes RS, Golde DW. Bone marrow origin of hepatic macrophages (Kupffer cells) in humans. Science. 1978;201:937–938. doi: 10.1126/science.356266. [DOI] [PubMed] [Google Scholar]
- Volc-Platzer B, Stingl G, Wolff K, Hinterberg W, Schnedl W. Cytogenetic identification of allogeneic epidermal Langerhans cells in a bone marrow graft recipient. N Engl J Med. 1984;310:1123–1124. doi: 10.1056/NEJM198404263101721. [DOI] [PubMed] [Google Scholar]
- Unger ER, Sung JH, Manivel JC, Chenggis ML, Blazar BR, Krivit W. Male donor-derived cells in the brains of female sex-mismatched bone marrow transplant recipients: a Y chromosome specific in situ hybridization study. J Neuropathol Exp Neurol. 1993;42:460–470. doi: 10.1097/00005072-199309000-00004. [DOI] [PubMed] [Google Scholar]
- Petersen BE, Bowen WC, Patrene KD, Mars WM, Sullivan AK, Murase N, Boggs SS, Greenberger JS, Goff JP. Bone marrow as a potential source of hepatic oval cells. Science. 1999;284:1168–1170. doi: 10.1126/science.284.5417.1168. [DOI] [PubMed] [Google Scholar]
- Theise ND, Badve S, Saxena R, Henegariu O, Sell S, Crawford JM, Krause DS. Derivation of hepatocytes from bone marrow cells in mice after radiation-induced myeloablation. Hepatology. 2000;31:235–240. doi: 10.1002/hep.510310135. [DOI] [PubMed] [Google Scholar]
- Krause DS, Theise ND, Collector MI, Henegariu O, Hwang S, Gardner R, Neutzel S, Sharkis SJ. Multi-organ, multi-lineage engraftment by a single bone marrow-derived stem cell. Cell. 2001;105:369–377. doi: 10.1016/s0092-8674(01)00328-2. [DOI] [PubMed] [Google Scholar]
- Lagasse E, Connors H, Al-Dhalimy M, Reitsma M, Dohse M, Osborne L, Wang X, Finegold M, Weissman IL, Grompe M. Purified hematopoietic stem cells can differentiate into hepatocytes in vivo. Nat Med. 2000;6:1229–1234. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- Poulsom R, Forbes SJ, Hodivala-Dilke K, Ryan E, Wyles S, Navaratnarasah S, Jeffery R, Hunt T, Alison M, Cook T, Pusey C, Wright NA. Bone marrow contributes to renal parenchymal turnover and regeneration. Pathology. 2001;195:229–235. doi: 10.1002/path.976. [DOI] [PubMed] [Google Scholar]
- Alison MR, Poulsom R, Jeffery R, Dhillon AP, Quaglia A, Jacob J, Novelli M, Prentice G, Williamson J, Wright NA. Hepatocytes from non-hepatic adult stem cells. Nature. 2000;406:257. doi: 10.1038/35018642. [DOI] [PubMed] [Google Scholar]
- Theise ND, Nimmakayalu M, Gardner R, Illei PB, Morgan G, Teperman L, Henegariu O, Krause DS. Liver from bone marrow in humans. Hepatology. 2000;32:11–16. doi: 10.1053/jhep.2000.9124. [DOI] [PubMed] [Google Scholar]
- Korbling M, Katz RL, Khanna A, Ruifrok AC, Rondon G, Albitar M, Champlin RE, Estrov Z. Hepatocytes and epithelial cells of donor origin in recipients of peripheral-blood stem cells. N Engl J Med. 2002;346:738–746. doi: 10.1056/NEJMoa3461002. [DOI] [PubMed] [Google Scholar]
- Okamoto R, Yajima T, Yamazaki M, Kanai T, Mukai M, Okamoto S, Ikeda Y, Hibi T, Inazawa J, Watanabe M. Damaged epithelia regenerated by bone marrow-derived cells in the human gastrointestinal tract. Nat Med. 2002;8:1011–1017. doi: 10.1038/nm755. [DOI] [PubMed] [Google Scholar]
- Abkowitz JL. Can human hematopoietic stem cells become skin, gut, or liver cells? N Engl J Med. 2002;346:770–772. doi: 10.1056/NEJM200203073461012. [DOI] [PubMed] [Google Scholar]
- DeWitt N, Knight J. Biologists question adult stem-cell versatility. Nature. 2002;416:354. doi: 10.1038/416354a. [DOI] [PubMed] [Google Scholar]
- Holden C, Vogel G. Stem cells. Plasticity: time for a reappraisal? Science. 2002;296:2126–2129. doi: 10.1126/science.296.5576.2126. [DOI] [PubMed] [Google Scholar]
- Blau HM, Brazelton TR, Weimann JM. The evolving concept of a stem cell: entity or function? Cell. 2001;105:829–841. doi: 10.1016/s0092-8674(01)00409-3. [DOI] [PubMed] [Google Scholar]
- Smallcombe A. Multicolor imaging: the important question of co-localization. BioTechniques. 2001;30:1240–1246. doi: 10.2144/01306bt01. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci USA. 1996;93:705–708. doi: 10.1073/pnas.93.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Paglieroni T, Ohto H, Holland PV, Busch MP. Survival of donor leukocyte subpopulations in immunocompetent transfusion recipients: frequent long-term microchimerism in severe trauma patients. Blood. 1999;93:3127–3139. [PubMed] [Google Scholar]
- Terada N, Hamazaki T, Oka M, Terada N, Hamazaki T, Oka M, Hoki M, Mastalerz DM, Nakano Y, Meyer EM, Morel L, Petersen BE, Scott EW. Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature. 2002;416:542–545. doi: 10.1038/nature730. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416:545–548. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- Wang X, Willenbring H, Akkari Y, Torimaru Y, Foster M, Al-Dhalimy M, Lagasse E, Finegold M, Olson S, Grompe M. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897–901. doi: 10.1038/nature01531. [DOI] [PubMed] [Google Scholar]
- Vassilopoulos G, Wang P, Russell DW. Transplanted bone marrow regenerates liver by cell fusion. Nature. 2003;422:901–904. doi: 10.1038/nature01539. [DOI] [PubMed] [Google Scholar]
- Tran S, Pillemer SR, Dutra A, Barett AJ, Brownstein MJ, Key S, Pak E, Leakan RA, Kingman A, Yamada KM, Baum BJ, Mezey E. Differentiation of human bone marrow-derived cells into buccal epithelial cells in vivo: a molecular analytical study. Lancet. 2003;361:1084–1088. doi: 10.1016/S0140-6736(03)12894-2. [DOI] [PubMed] [Google Scholar]
- Quesenberry PJ, Colvin GA, Lambert JF. The chiaroscuro stem cell: a unified stem cell theory. Blood. 2002;100:4266–4271. doi: 10.1182/blood-2002-04-1246. [DOI] [PubMed] [Google Scholar]
- Artlett CM, Smith JB, Jimenez SA. Identification of fetal DNA and cells in skin lesions from women with systemic sclerosis. N Engl J Med. 1998;338:1186–1191. doi: 10.1056/NEJM199804233381704. [DOI] [PubMed] [Google Scholar]
- Srivatsa B, Srivatsa S, Johnson KL, Samura O, Lee SL, Bianchi DW. Microchimerism of presumed fetal origin in thyroid specimens from women: a case-control study. Lancet. 2001;358:2034–2038. doi: 10.1016/S0140-6736(01)07099-4. [DOI] [PubMed] [Google Scholar]
- Jones DE. Fetal microchimerism: an aetiological factor in primary biliary cirrhosis? J Hepatol. 2000;33:834–837. doi: 10.1016/s0168-8278(00)80317-7. [DOI] [PubMed] [Google Scholar]
- Lagaaij EL, Cramer-Knijnenburg GF, van Kemenade FJ, van Es LA, Bruijn JA, van Krieken JH. Endothelial cell chimerism after renal transplantation and vascular rejection. Lancet. 2001;357:33–37. doi: 10.1016/S0140-6736(00)03569-8. [DOI] [PubMed] [Google Scholar]
- Hess D, Li L, Martin M, Sakano S, Hill D, Strutt B, Thyssen S, Gray DA, Bhatia M. Bone marrow-derived stem cells initiate pancreatic regeneration. Nat Biotechnol. 2003;21:763–770. doi: 10.1038/nbt841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.