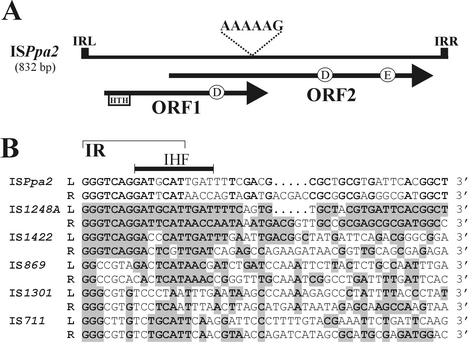
FIG. 3.
(A) Genetic organization of ISPpa2. IRLs and IRRs are shown as solid boxes. The two consecutive overlapping ORFs (ORF1 and ORF2) and their transcriptional orientation are shown by arrows. The location of the DNA binding domain (HTH) of ORF1 and the DDE motif of a putative fusion protein are marked. The location of a putative frameshifting motif (5′-AAAAAG-3′) is indicated. (B) Alignment of the terminal nucleotide sequences of ISPpa2 and its closest relatives of the IS427 group (IS5 family). The identical residues of termini of ISPpa2 are boldface. Nucleotides of other sequences identical with those of ISPpa2 are boldface and shaded. A putative site for binding IHF, present in the IRL of ISPpa2, is indicated by a thick bar. The putative IRs of ISPpa2 are depicted by a thin bar. “L” denotes sequences at the 5′ (left) end, and “R” denotes complementary sequences at the 3′ (right) end of the elements.