Abstract
The mukB gene is essential for the partitioning of sister chromosomes in Escherichia coli. A mukB null mutant is hypersensitive to the DNA gyrase inhibitor novobiocin. In this work, we isolated mutants suppressing the novobiocin hypersensitivity of the mukB null mutation. All suppressor mutations are localized in or near the gyrB gene, and the four tested clones have an amino acid substitution in the DNA gyrase beta subunit. We found that in the mukB mutant, the process of sister chromosome segregation is strikingly hypersensitive to novobiocin; however, the effect of novobiocin on growth, which was measured by culture turbidity, is the same as that of the wild-type strain.
The smtA-mukF-mukE-mukB operon (22) is located at the 21-min map position of the Escherichia coli chromosome. Deletion mutants of each muk gene show common phenotypes, such as frequent production of anucleate cells upon cell division, chromosome guillotining (cutting) by septum closure, and temperature-sensitive growth; however, they are nearly normal in chromosome replication, homologous recombination, mutation frequency, and sensitivity to UV irradiation (10, 11, 22; for a review, see reference 4). MukF, MukE, and MukB form a large complex (22). MukB is a member of the SMC superfamily (9). A MukB-green fluorescent protein fusion protein is localized as one, two, or four foci at regular cellular positions in the presence of both MukF and MukE (12). To examine the function of the MukB protein, many suppressor mutants of mukB mutations have been analyzed (for a review, see reference 4). Mutations of the topA gene encoding topoisomerase I suppress temperature-sensitive growth and anucleate-cell production (16). It has been demonstrated in synchronized cultures that replicated sister chromosomes are associated with one another for substantial times (6, 18). In contrast, results in randomly growing cultures in enriched medium appear to be consistent with the model in which replicated sister copies of oriC separate immediately after replication (8). However, it is hard to determine which model is right from the results of such random cultures, because the actual time of initiation of chromosome replication is not clear in this type of experiment.
A mukB null mutant is hypersensitive to novobiocin, an inhibitor (20). Deletion mutants of each of the mukB, mukF, and mukE genes and a deletion mutant lacking all three muk genes are hypersensitive to novobiocin (14). Weitao et al. (20) showed that a seqA null mutation suppressed temperature-sensitive growth, anucleate-cell production, and novobiocin hypersensitivity in the mukB null mutation. Inconsistently, Onogi et al. (14) reported that a seqA or dam null mutation partially suppressed temperature-sensitive growth but failed to suppress the anucleate-cell production and novobiocin hypersensitivity of these muk null mutants.
It is not yet clear what the mechanism of the novobiocin hypersensitivity of these muk mutants is. What is the target protein that is hypersensitive to novobiocin in these muk null mutants? There are two possibilities. First, DNA gyrase could be the target of a low concentration of novobiocin in muk null mutants. Second, an unknown protein could be the target, which should be more sensitive to novobiocin than DNA gyrase and essential for growth only in the defective muk background but nonessential in the wild-type muk genetic background. In this work, to investigate the identity of the target protein, we isolated novobiocin-resistant suppressor mutants from the mukB null mutant strain and characterized them. We found that the beta subunit of DNA gyrase is the target protein of novobiocin in a mukB null mutant. We discuss the mechanism of novobiocin hypersensitivity of mukB null mutant cells.
MATERIALS AND METHODS
Bacterial strains.
YK1100 (trpC9941) and AZ5372 (trpC9941 ΔmukB::kan) are derivatives of E. coli strain W3100, as described previously (14, 22). Bacterial cells were grown in Luria-Bertani (LB) medium at 22°C, which is the permissive temperature for AZ5372. QT186 (strain JC12334 [15]) has the tna-300::Tn10 marker, which is linked with the gyrB gene.
Isolation of mutants that suppress novobiocin hypersensitivity of a mukB null mutation.
Ten independent single colonies of strain AZ5372 grown on LB agar plates at 22°C were inoculated into LB liquid medium, and the cultures were grown to saturation at 22°C. Fifty microliters of each culture was spread onto an LB agar plate containing 100 or 1,000 μg of novobiocin/ml and incubated at 22°C for 5 days. Confluent growth of cells occurred on the plates containing 100 μg of novobiocin/ml, due to the large number of cells spread on the plates. However, only 10 to 30 novobiocin-resistant colonies appeared on each plate containing 1,000 μg of novobiocin/ml. Single colonies from each plate were picked and purified on LB agar plates at 22°C and named MQ40, MQ41, and MQ43 to MQ50. These purified clones were shown to have the same tryptophan requirement and kanamycin resistance at 22°C as the parental strain, AZ5372.
Sensitivity to DNA gyrase inhibitors.
The colony-forming abilities of various strains were analyzed on LB agar plates containing various concentrations of novobiocin or nalidixic acid according to the method of Onogi et al. (14). Novobiocin and nalidixic acid were obtained from Sigma Chemical Co.
Transduction with phage P1vir.
Suppressor mutant cells were infected with phage P1vir propagated on QT186 cells. About 500 tetracycline-resistant transductants were obtained on each LB agar plate containing 7.5 μg of tetracycline/ml after incubation at 22°C for 6 days. Fifty of the transductants were isolated from each plate, purified, and analyzed for novobiocin hypersensitivity on LB agar plates containing 300 μg of novobiocin/ml after incubation for 5 days at 22°C.
DNA sequencing.
To determine the mutation sites by DNA sequencing, we amplified the 2,580 bp of DNA fragments comprising the gyrB gene and its flanking regions from chromosomal DNA samples of four suppressor mutants (MQ40, MQ43, MQ44, and MQ45) and the parental AZ5372 strain by PCR using the forward primer 5′GTGCTGAACACGTTATAGACATGTCGGACG3′ and the reverse primer 5′CAAGATTTTCGTAGGCCTGATAAGCGTAGC3′.
Fluorescence microscopy.
Cells were stained with DAPI (4′,6-diamino-2-phenylindole) for chromosomal DNA and observed with fluorescence and phase-contrast microscopes according to the method of Hiraga et al. (5).
Survival after irradiation with X rays.
Cells grown exponentially in LB medium at 22°C were irradiated by X rays, diluted, spread on LB agar plates, and incubated at 22°C for 5 days. The colonies that appeared were counted.
RESULTS
Isolation and characterization of mutants that suppress novobiocin hypersensitivity of a mukB null mutation.
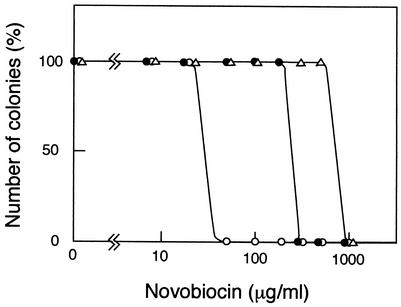
Cells grown at 22°C in LB medium were diluted, spread on LB agar plates containing various concentrations of novobiocin, and incubated at 22°C for 5 days. As shown in Fig. 1, the wild-type strain YK1100 (a trpC9941 derivative of strain W3110) was able to form colonies in the presence of 200 μg of novobiocin/ml but not at 300 μg/ml. In contrast, the isogenic mukB null mutant AZ5372 (W3110 trpC9941 ΔmukB::kan) was able to form colonies at 20 μg/ml but not at 50 μg/ml. Thus, the maximum concentrations of novobiocin allowing the survival of >50% of cells were 200 μg/ml in YK1100 and 20 μg/ml in AZ5372. The mukB null mutant was 10-fold more sensitive to novobiocin than the wild-type strain, consistent with previous results (14).
FIG. 1.
Novobiocin sensitivities of various strains in colony formation. Solid circles, parental strain YK1100 (W3110 trpC9941); open circles, AZ5372 (W3110 trpC9941 ΔmukB::kan); open triangles, novobiocin-resistant suppressor mutants that were independently isolated from AZ5372. All 10 suppressor mutants showed the same sensitivity to novobiocin.
To identify the target of novobiocin in the mukB null mutant, we isolated 10 novobiocin-resistant mutants in which the novobiocin hypersensitivity of the mukB null mutation was suppressed. These suppressor clones were analyzed for novobiocin sensitivity, temperature sensitivity, and anucleate-cell formation. All 10 clones were able to grow at 22°C in the presence of 500 μg of novobiocin/ml but not at 1,000 μg/ml (Fig. 1 and Table 1). Two clones, MQ40 and MQ41, showed partial suppression of temperature sensitivity; colony-forming abilities at 37°C were 6.5 × 10−3 and 1.2 × 10−2, respectively, compared with that at 22°C. The other eight clones showed the same temperature sensitivity as the parental mukB null mutant; colony-forming ability at 37°C was <2 × 10−6 compared with that at 22°C (Table 1). All novobiocin-resistant clones retained the property of frequent production of anucleate cells at 22°C.
TABLE 1.
Properties of 10 mutants suppressing novobiocin hypersensitivity of mukB null mutation
| Strain | Growth at 22°C with indicated concn of novobiocin (μg/ml)a
|
Colony-forming ability atb:
|
Formation of anucleate cellsc | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 20 | 100 | 300 | 500 | 22°C | 37°C | ||
| YK1100 | + | + | + | − | − | 1 | 1.3 | − |
| AZ5372 | + | + | − | − | − | 1 | <2 × 10−6 | ++ |
| MQ40 | + | + | + | + | + | 1 | 6.5 × 10−3 | ++ |
| MQ41 | + | + | + | + | + | 1 | 1.2 × 10−2 | ++ |
| MQ43 | + | + | + | + | + | 1 | <2 × 10−6 | ++ |
| MQ44 | + | + | + | + | + | 1 | <2 × 10−6 | ++ |
| MQ45 | + | + | + | + | + | 1 | <2 × 10−6 | ++ |
| MQ46 | + | + | + | + | + | 1 | <2 × 10−6 | ++ |
| MQ47 | + | + | + | + | + | 1 | <2 × 10−6 | ++ |
| MQ48 | + | + | + | + | + | 1 | <2 × 10−6 | ++ |
| MQ49 | + | + | + | + | + | 1 | <2 × 10−6 | ++ |
| MQ50 | + | + | + | + | + | 1 | <2 × 10−6 | ++ |
+, growth; −, no growth.
Colony-forming ability on LB agar medium at 22°C was defined as 1.
−, no anucleate cell; ++, many anucleate cells.
Mapping of suppressor mutations.
To map the suppressor mutations, we first determined whether they are located in the gyrB gene, which encodes the beta subunit of DNA gyrase. The wild-type gyrB gene, located at the 83-min map position, can be cotransduced by phage P1vir together with the tna-300::Tn10 marker of strain QT186, which is located near the gyrB gene. Eighty to 95% of tetracycline-resistant transductants in each case exhibited the same degree of novobiocin hypersensitivity as the parent strain, AZ5372. This suggested that when the wild-type gyrB gene of the donor QT186 was cotransduced with the tna-300::Tn10 marker, the transductants became hypersensitive to novobiocin, like the parental AZ5372. This implies that the suppressor mutations are located in or near the gyrB gene. MQ40 and MQ41 showed partial suppression of temperature sensitivity as described above (Table 1). Novobiocin-hypersensitive clones among tetracycline-resistant transductants from MQ40 and MQ41were unable to form colonies at 37°C on LB agar plates, like the parental AZ5372, implying that the mutation that partially suppresses temperature sensitivity is closely linked to the mutation conferring novobiocin resistance. To avoid the possibility of contamination, all tetracycline-resistant transformants were confirmed to have a tryptophan requirement and kanamycin resistance, like the parental strain, AZ5372.
We sequenced the gyrB DNA fragments obtained from MQ40, MQ43, MQ44, MQ45, and AZ5372. We found that in all suppressor mutants tested, the guanosine residue at position 407 (the adenosine residue of the start codon ATG is defined as +1) is changed to thymidine, resulting in the replacement of arginine at position 136 by leucine of the beta subunit of DNA gyrase. The arginine residue at position 136 is conserved in the GyrB proteins of various bacterial species. This arginine residue is one of the key amino acids implicated in novobiocin binding (7; http://www.sanger.ac.uk/Software/Pfam/index.shtml). MQ40 showed weak suppression of temperature sensitivity, in contrast to the other sequenced clones (Table 1). We speculate that MQ40 has a second mutation, which affects expression of the altered GyrB protein, outside the sequenced region. The gyrB gene is known to be located in an operon, dnaA-dnaN-recF-gyrB.
Effect of novobiocin on cell growth and chromosome segregation in mukB null mutant cells.
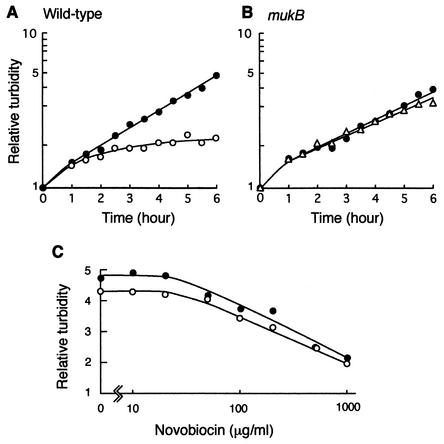
To examine why the mukB null mutant is hypersensitive to low concentrations of novobiocin, we analyzed the effect of novobiocin on increase of turbidity of cultures in wild-type and mukB mutant strains. Cultures growing in LB medium at 22°C were incubated for 6 h in the presence of cephalexin alone or cephalexin plus novobiocin, as shown in Fig. 2. Cephalexin (10 μg/ml) was added to the cultures in order to inhibit cell division to prevent guillotining of the chromosome by septum closure in the mukB null mutant (4, 11, 22), because we expected that the abnormal structure of the chromosome in mukB mutant cells would be clearly observed in elongated cells incubated in the presence of cephalexin. Novobiocin was added at concentrations of 1,000 μg/ml in YK1100 and 100 μg/ml in AZ5372. These concentrations were five-fold higher than the maximum concentration of novobiocin allowing the survival of >50% of cells in each strain (Fig. 1). When cephalexin (10 μg/ml) alone was added, the turbidity of cultures increased exponentially for at least 6 h in both strains, suggesting that DNA and protein syntheses continued normally, although cell division was inhibited by cephalexin. The doubling times measured by turbidity increase were 155 and 190 min in YK1100 and AZ5372, respectively, under the conditions used. When 1,000 μg of novobiocin/ml was added to the culture of YK1100 together with cephalexin, the turbidity increased ∼1.5-fold for the first 2 h; however, the increase in turbidity was markedly inhibited after that (Fig. 2A). On the other hand, surprisingly, when 100 μg of novobiocin/ml was added to the culture of AZ5372, the turbidity increased exponentially for 6 h without any significant inhibitory effect (Fig. 2B), although the mukB mutant was unable to form colonies in the presence of the same concentration of novobiocin (Fig. 1). To confirm these results, we analyzed the effects of various concentrations of novobiocin on increase of turbidity after 6 h of incubation in both strains. As shown in Fig. 2C, the effects of novobiocin on growth were the same in both strains, implying that the effects of novobiocin on DNA and protein syntheses were the same in both strains. This indicates that the effect of muk mutation on the structure of chromosomal DNA, for example, decompaction or reduced superhelicity of chromosomal DNA (16, 19-21), does not affect DNA and protein syntheses, consistent with expectations. Thus, mukB mutant cells lost colony-forming ability in the presence of a low concentration of novobiocin, such as 100 μg/ml, even though DNA and protein syntheses continued at the levels of the wild-type strain. One might ask what the mechanism of novobiocin hypersensitivity is in the mukB mutant.
FIG. 2.
Effects of novibiocin on growth of cultures in YK1100 and AZ5372. (A) YK1100. Solid circles, cephalexin (10 μg/ml) alone; open circles, cephalexin plus 1,000 μg of novobiocin/ml. (B) AZ5372. Solid circles, cephalexin alone; open triangles, cephalexin plus 100 μg of novobiocin/ml. (C) Final value of turbidity after incubation for 6 h with or without indicated concentrations of novobiocin. The turbidity at zero time is 1. Solid circles, YK1100; open circles, AZ5372.
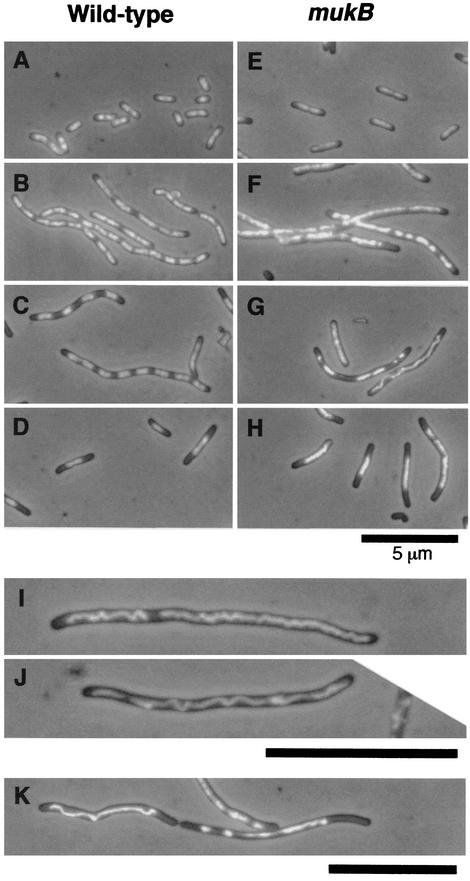
To answer this question, we observed cells by fluorescence microscopy after a 6-h incubation in the presence of cephalexin and novobiocin. We found a striking effect on the structure of nucleoids in mukB mutant cells at 100 μg of novobiocin/ml. The nucleoids were large and frequently formed long twisted strings in elongated filamentous cells of AZ5372, as shown in Fig. 3G, I, J, and K. In contrast, nucleoids were normal in size and arranged at nearly equal distances in elongated filamentous cells in YK1100 at the same concentration of novobiocin (Fig. 3C). These results indicate that segregation of sister chromosomes is strongly inhibited by a low concentration of novobiocin in the mukB mutant but not in the wild-type strain.
FIG. 3.
Merged images of cells stained with DAPI by fluorescence and phase-contrast microscopy. (A to D) YK1100; (E to K) AZ5372. (A and E) Before the addition of antibiotics. (B and F) Cephalexin (10 μg/ml) alone for 6 h. (C, G, I, J, and K) Cephalexin (10 μg/ml) and novobiocin (100 μg/ml) for 6 h. (D and H) Cephalexin (10 μg/ml) and novobiocin (1,000 μg/ml) for 6 h. The scale bars represent 5 μm.
After incubation with 1,000 μg of novobiocin/ml, YK1100 cells were small and had one nucleoid that was localized in the center of the cell or at the constriction site at midcell (Fig. 3D). In a minority of YK1100 cells, two nucleoids existed on both sides, flanking the constriction site. These results suggest strong inhibition of DNA and protein syntheses under these conditions, as expected. In AZ5372 cells incubated with 1,000 μg of novobiocin/ml, nucleoids were frequently larger than those of YK1100 cells (Fig. 3H). The large size of nucleoids in mukB mutant cells is presumably a result of a defect in the segregation of sister chromosomes (2, 6; M. Kohiyama, T. Onogi, S. Adachi, and S. Hiraga, unpublished data) before and after the addition of antibiotics.
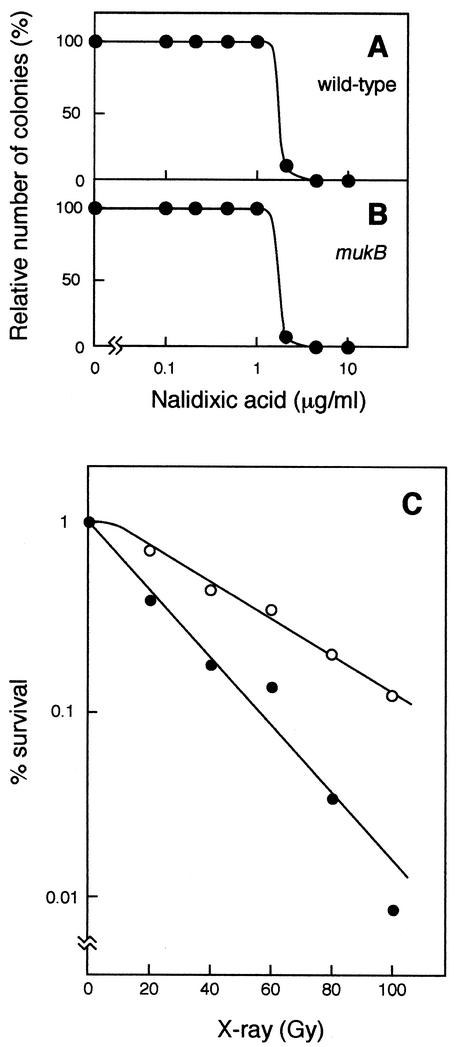
Sensitivity to nalidixic acid and X-ray irradiation in mukB null mutant cells.
We analyzed mukB null mutant cells for sensitivity to another DNA gyrase inhibitor, nalidixic acid. The maximum concentration of nalidixic acid allowing colony formation was 1 μg/ml in both wild-type and mukB null mutant strains (Fig. 4A and B). The mukB mutant showed more resistance to X-ray irradiation than the wild-type strain (Fig. 4C). These results are discussed below.
FIG. 4.
Effects of nalidixic acid on colony-forming ability (A and B) and survival after X-ray irradiation (C). (A) YK1100; (B) AZ5372; (C) YK1100 (solid circles) and AZ5372 (open circles).
DISCUSSION
The above-mentioned results indicate that the target protein that is hypersensitive to novobiocin in the mukB null mutant is DNA gyrase. Novobiocin inhibits DNA gyrase, resulting in a reduction in the superhelicity of circular DNA (1, 3). Another DNA gyrase inhibitor, nalidixic acid, inhibits DNA gyrase and produces double-strand breaks in DNA (17). We have observed no significant difference in sensitivity to nalidixic acid between the wild-type and mukB null mutant strains (Fig. 4A and B). This is consistent with our observation that the mukB mutant is more resistant to X-ray irradiation than the wild-type strain (Fig. 4C). The resistance to X rays might be due to a larger average number of chromosomes in mukB mutant cells (Kohiyama et al., unpublished). X-ray irradiation also induces double-strand breaks in DNA (2). These results indicate that the mukB null mutant is hypersusceptible to the reduction in superhelicity of chromosomal DNA but not to double-strand breaks. In this work, we found no significant difference between wild-type and mukB mutant strains in the novobiocin concentration that inhibits growth of cultures as measured by turbidity increase (Fig. 2). However, segregation of sister chromosomes is remarkably inhibited in AZ5372, but not in YK1100, by a low concentration (100 μg/ml) of novobiocin (Fig. 3). The abnormality in segregation of sister chromosomes should cause lethality (inability to form colonies) in mukB mutant cells. A step in the process of sister chromosome segregation might depend strongly on the DNA supercoiling function catalyzed by DNA gyrase in the mukB null mutant at the permissive temperature, 22°C. In mukB null mutant cells, the abnormal segregation in the presence of a low concentration of novobiocin might cause an irreversible defect, resulting in the loss of colony-forming ability.
In wild-type cells, the replisome complexes associated with diverging replication forks are closely associated with one another at midcell during the early phase of replication; however, they separate and migrate rapidly to one-quarter and three-quarter cell positions during replication. This event causes persistent separation of clockwise and counterclockwise replicating regions of the chromosome (4, 6, 13, 18). Protein-protein linkage between clockwise and counterclockwise replicating regions, e.g., by the hemimethylated DNA binding SeqA protein, might be broken off by the event. Two clusters of the MukFEB complex known to be located at the cell quarter positions (12) might participate in the event of migration of replication forks, because the mukB mutant has a defect in the regular subcellular localization of replication forks (Kohiyama et al., unpublished). Flow cytometry and immunofluorescence microscopy revealed that separation of sister chromosomes is delayed in mukB null mutant cells (Kohiyama et al., unpublished). This is consistent with frequent production of anucleate cells upon cell division in the mutant. The MukFEB complex might facilitate the resolution of interwound sister chromosomal DNA strands to form two separated nucleoids. Reduced superhelicity of bacterial and plasmid DNAs was observed after proteins in the mukB null mutant were removed (16, 19-21). However, no significant difference between the levels of compactness of nascent DNA labeled with 5-bromodeoxyuridine and bulk chromosomal DNA could be detected in the wild-type and mukB null mutant strains by immunofluorescence microscopy and fluorescence microscopy in vivo (Kohiyama et al., unpublished), in contrast to in vitro data (19-21). SeqA participates in the compactness of nascent and bulk chromosomal DNA in vivo, because decompaction of the chromosome was observed in the seqA null mutant but not in the dam null mutant in vivo (Kohiyama et al., unpublished). Temperature-sensitive growth, but not novobiocin hypersensitivity, of the mukB null mutation was suppressed by the seqA or dam null mutation (14). These results suggest that linkages of SeqA-SeqA interaction between nascent DNA segments in clockwise and counterclockwise replicating regions are probably harmful for cell growth in the genetic background lacking the MukFEB complex above 30°C in rich media. The MukFEB complex would act for separation of the SeqA-SeqA linkages between nascent DNA segments of clockwise and counterclockwise replicating regions. This idea is consistent with the phenomenon that the seqA or dam null mutations suppress the temperature sensitivity of the mukB null mutation.
Acknowledgments
We thank Akiko Nishimura for bacterial strains.
S. Hiraga was supported by a grant from the Center of Excellence (COE).
REFERENCES
- 1.Ali, J. A., A. P. Lackson, A. J. Howells, and A. Maxwell. 1993. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry 32:2717-2724. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 3.Gellert, M., M. H. O'Dea, T. Itoh, and J. Tomozawa. 1976. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. USA 73:4474-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiraga, S. 2000. Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet. 34:21-59. [DOI] [PubMed] [Google Scholar]
- 5.Hiraga, S., H. Niki, T. Ogura, C. Ichinose, H. Mori, B. Ezaki, and A. Jaffé. 1989. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J. Bacteriol. 171:1496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiraga, S., C. Ichinose, T. Onogi, H. Niki, and M. Yamazoe. 2000. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells 5:327-341. [DOI] [PubMed] [Google Scholar]
- 7.Lewis, R. J., O. M. P. Singh, C. V. Smith, T. Skarzynski, A. Maxwell, A. J. Wonacott, and D. B. Wigley. 1996. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 15:1412-1420. [PMC free article] [PubMed] [Google Scholar]
- 8.Li, Y., K. Sergueev, and S. Austin. 2002. The segregation of the Escherichia coli origin and terminus of replication. Mol. Microbiol. 46:985-995. [DOI] [PubMed] [Google Scholar]
- 9.Melby, T. E., C. N. Ciampaglio, G. Briscore, and H. P. Erickson. 1998. The symmetrical structure maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 142:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niki, H., R. Imamura, M. Kitaoka, K. Yamanaka, T. Ogura, and S. Hiraga. 1992. E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 11:5101-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niki, H., A. Jaffé, R. Imamura, T. Ogura, and S. Hiraga. 1991. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10:183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohsumi, K., M. Yamazoe, and S. Hiraga. 2001. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol. Mirobiol. 40:835-845. [DOI] [PubMed] [Google Scholar]
- 13.Onogi, T., K. Ohsumi, T. Katayama, and S. Hiraga. 2002. Replication-dependent recruitment of the β-subunit of DNA polymerase III from cytosolic spaces to replication forks in Escherichia coli. J. Bacteriol. 184:867-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onogi, T., M. Yamazoe, C. Ichinose, H. Niki, and S. Hiraga. 2000. Null mutation of the dam or seqA gene suppresses temperature-sensitive lethality but not hypersensitivity to novobiocin of muk null mutants. J. Bacteriol. 182:5898-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ream, L. W., L. Margossian, and J. C. Alvin. 1980. Genetic and physical mapping of recF in Escherichia coli K-12. Mol. Gen. Genet. 180:115-121. [DOI] [PubMed] [Google Scholar]
- 16.Sawitzke, J. A., and S. Austin. 2000. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl. Acad. Sci. USA 97:1671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugino, A., C. L. Peebles, K. N. K. Kreuzer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunako, Y., T. Onogi, and S. Hiraga. 2001. Sister chromosome cohesion of Escherchia coli. Mol. Microbiol. 42:1233-1241. [DOI] [PubMed] [Google Scholar]
- 19.Weitao, T., S. Dasgupta, and K. Nordström. 2000. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol. Microbiol. 38:392-400. [DOI] [PubMed] [Google Scholar]
- 20.Weitao, T., K. Nordström, and S. Dasgupta. 1999. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol. 34:157-168. [DOI] [PubMed] [Google Scholar]
- 21.Weitao, T., K. Nordström, and S. Dasgupta. 2000. Escherichia coli cell cycle control genes affect chromosome superhelicity. EMBO Rep. 1:494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamanaka, K., T. Ogura, H. Niki, and S. Hiraga. 1996. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol. Gen. Genet. 250:241-251. [DOI] [PubMed] [Google Scholar]
- 23.Yamazoe, M., T. Onogi, Y. Sunako, H. Niki, K. Yamanaka, T. Ichimura, and S. Hiraga. 1999. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 18:5873-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]