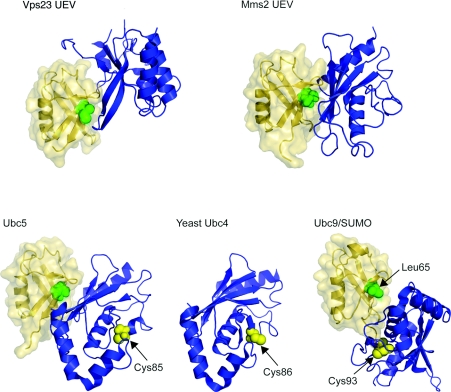
Figure 6. Ubc-related domain structures.
Upper panel: UEV domains of Vps23 and Mms2 are shown (blue) in ribbon representation with ubiquitin (yellow) in ribbon and surface representations. Ile44, the centre of the hydrophobic recognition patch on the ubiquitin, is shown as green spheres. Ubiquitin molecules are placed in the same orientation as in Figure 1 for comparison. Protein data bank identication codes used are: Vps23 UEV, 1UZX; Mms2 UEV, 1ZGU. Lower panels: Ubc5 (blue) is shown on the left with ubiquitin (yellow). The catalytically important Cys85 is depicted as yellow sphere. Yeast Ubc4 is presented in the middle panel for comparison. Again, catalytic Cys86 is shown as yellow sphere. On the right, Ubc9 (blue) complexed with SUMO (small ubiquitin-related modifier) (yellow) is shown [109], as there is no equivalent structure of a Ubc–ubiquitin conjugate available. Leu65 on SUMO, equivalent to Ile44 from sequence alignment, is indicated as green sphere and Cys93 of Ubc9 as yellow sphere. Protein data bank identication codes used are: Ubc5, 2FUH; Ubc4, 1QCQ; Ubc9/SUMO, 1Z5S. The SUMO molecule is also placed in the same orientation as the ubiqutin molecule.