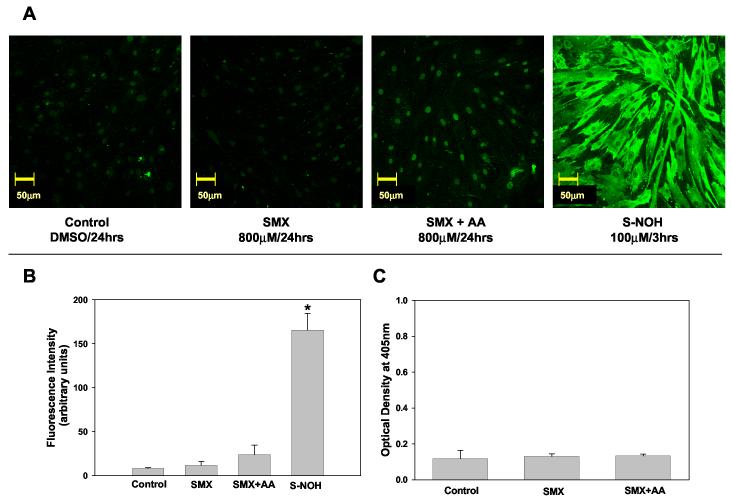
Figure 1.
Sulfamethoxazole (SMX) - protein adducts formation in NHDF.
A) Detection of SMX-protein adducts using confocal microscopy. NHDF cells (5x104 cells/mL) were incubated with SMX (800 μM, 24 h) in presence or absence of ascorbic acid (2 mM) or S-NOH (100 μM, 3 h). Controls were exposed to the vehicle alone (dimethyl sulfoxide). At the end of the incubation cells were fixed with paraformaldehyde, permeabilized, immunostained, and imaged on a confocal microscope using 20X objective. B) Confocal Images were analyzed as detailed in the Materials and Methods and fluorescence intensity from a minimum of 3 view fields of 4 different slides (incubations) of each treatment (with 10 cells per field) were averaged and expressed as mean (+SD) fluorescence intensity (arbitrary units). Hence, each bar represents mean (SD) of the mean of 4 incubations with 30 measurements of fluorescence intensity for each incubation. Results were analyzed using ANOVA with Holm-Sidak method for multiple pairwise comparisons.*p<0.05 compared to control/SMX/SMX+AA. C) Detection of SMX-protein adducts using ELISA technique. NHDF cells from the same individual as in A & B were seeded at a density of 2x105 cells/mL and incubated in the presence or absence of SMX 800 μM and/or ascorbic acid (AA) 2 mM. Data presented as optical density (OD) mean (+SD) of 4 separate incubations for each condition. Results were analyzed using ANOVA with Holm-Sidak method for multiple pairwise comparisons. There is no statistical difference (p<0.05) between the treatments.