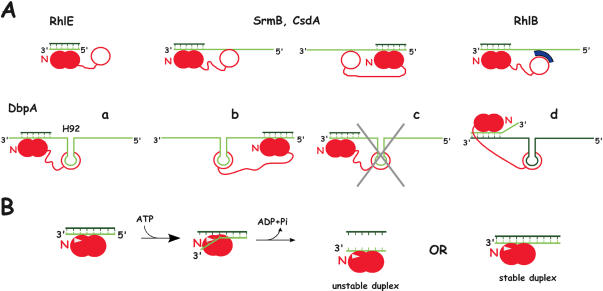
Figure 2.
Models for the unwinding activity of E.coli DEAD-box proteins. (A) Substrate recognition. The helicase is shown in red, with the core RecA-like domains and the C-terminal domain appearing as closed and open circles, respectively. The helicase core is assumed to bind to one strand of the RNA duplex, as observed with the DEAD-box protein Vasa in the presence of ATP (6); this strand is shown in light green, with the complementary strand in dark green. Moreover, the relative orientations of the core and bound RNA are assumed to be the same as in Vasa, i.e. the N-terminal domain (N) interacts with the 3′ side of the RNA. For the sake of simplicity, putative secondary binding sites (SrmB, CsdA, RhlB) are assumed to bind the same strand as the core, although this is not necessarily the case [cf DbpA, d; (23)]. (i) RhlE can unwind blunt end duplexes, and therefore its core presumably interacts directly with one strand of the duplex. (ii) SrmB and CsdA require single-stranded extensions, but these extensions can be either 5′ (left) or 3′ (right) to the duplex. We propose that in this case also the core interacts directly with one strand of the duplex, and that the extensions are used not for translocation but for binding the helicase via a secondary RNA binding site, which, by analogy with DbpA (see below), is shown here within the C-terminal domain. (iii) RhlB is inactive as a helicase unless it binds RNase E or a fragment thereof (in blue) carrying an arginine-rich region. This fragment may constitute a secondary RNA binding site that interacts with single-stranded extensions, as proposed above. The RhlB region that contacts RNase E has not been determined; it is assumed here to lie in the C-terminal domain. (iv) The DbpA C-terminal domain binds tightly to helix 92 (H92) of 23S rRNA, and the protein can unwind duplexes that are located either 3′ (a) or 5′ (b) to the helix. DbpA is unique amongst E.coli DEAD-box helicases in strictly requiring a 3′ single-stranded extension: substrates lacking such extension cannot be unwound (c). However, this extension does not need to lie on the same strand as helix 92 (d). (B) Model explaining why most DEAD-box helicases are poorly processive and do not require a single-stranded extension of definite polarity [based on (6)]. In the presence of ATP, the protein binds tightly to RNA, which can be either single- or double-stranded. However, the presence of a ‘wedge’ (white triangle) encompassing motif Ib (Figure 1) induces a kink in the bound RNA that is locally incompatible with double strandedness. Therefore, bound duplexes unwind over a few bases. The protein may preferentially bind near the duplex ends so that thermal fraying assists unwinding. After ATP hydrolysis, the partially unwound duplex is free to dissociate (‘unstable duplex’) or reanneal (‘stable duplex’), depending upon its stability. At this stage, the protein may either dissociate, or remain RNA-bound as illustrated here.