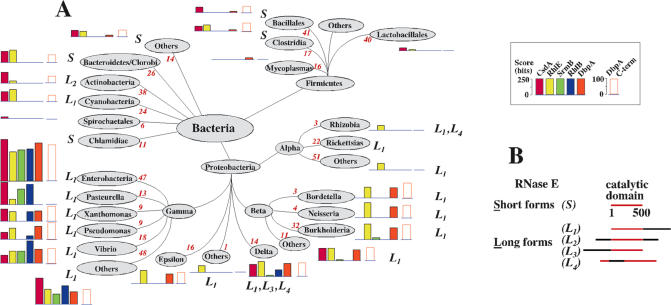
Figure 3.
(A) Distribution of putative orthologues of the E.coli DEAD-box proteins over the bacterial phylogenic tree, as revealed by BLAST analysis. The whole sequence of the proteins (coloured rectangles) or, for DbpA, the C-terminal extension (open rectangles; see Figure 1 and text for details) have been used as probes. The tree was adapted from a web page of The National Center for Biotechnology Information, National Library of Medicine, USA (www.ncbi.nlm.nih.gov). For a given DEAD-box protein (see inset for the colour code) and for each phylum, class or order, the height of the histogram is proportional to the highest score observed for a representative of this phylum, class or order. The scores obtained with the whole sequence probes have been corrected to take into account the homology that exists between unrelated DEAD-box proteins (see text). Red figures (italics) correspond to the number of sequences in the databank. Note that eventually bacteria distant from E.coli contain other DEAD-box helicases whose sequence are unrelated to those of E.coli and therefore have not been detected here. The mention ‘S’ or ‘L’ following the histogram refers to RNase E, the genuine partner of RhlB in E.coli. ‘S’ (Short) means that all identified RNase E orthologues consist of the catalytic region only, whereas ‘L’ (Long) means that at least one RNase E representative carries an extension of >100 residues with no homology to the catalytic core of E.coli RNase E (first 500 residues). Such extension could act as a scaffold for the assembly of the degradosome. The subscript (1–4) refers to the position of the extension with respect to the catalytic core (see B). (B) Schematic drawing showing the different configurations of RNase E observed in the phylogenetic tree.