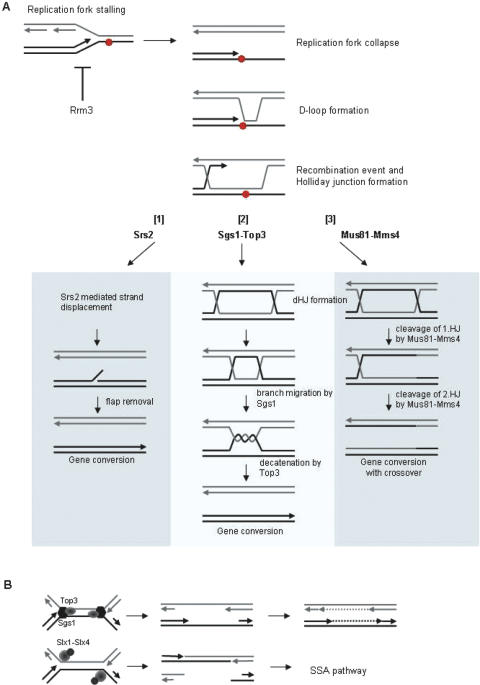
Figure 3.
Pathways where Sgs1–Top3 function to process collapsed or stalled replication forks. (A) When a replication fork approaches a lesion (red circle) in the DNA a single-stranded gap forms and replication fork collapse may occur. This allows Rad51-dependent D-loop formation to initiate a recombination event via Holliday Junction (HJ) formation. The HJ may be processed in three ways. [1] The invading strand can be disrupted by the Srs2 helicase as part of the SDSA pathway leading to gene conversion without crossover events. [2] The Sgs1–Top3 complex can work in a dissolution pathway, where Sgs1 first promotes branch migration on the formed dHJ substrate. The single-stranded interlinked DNA is subsequent decatenated by Top3 resulting in gene conversion without crossover events. [3] Finally endonucleases such as Mus81–Mms4 may process the dHJ by two subsequent cleavage reactions. This will generate gene conversion products either with, or without an associated crossover event. Note that in the absence of Rrm3 more stalled replication forks will be generated. (B) When replication forks converge and stall at the rDNA, Sgs1–Top3 may allow DNA replication to finish by a DNA polymerase fill in reaction after separating the parental strands of the DNA template. In the absence of Sgs1–Top3, Slx1–Slx4 may cleave at each replication fork thereby creating a double nicked chromatid and a broken sister chromatid. The DSB can be repaired by a RAD52-independent single-strand annealing pathway at the rDNA.