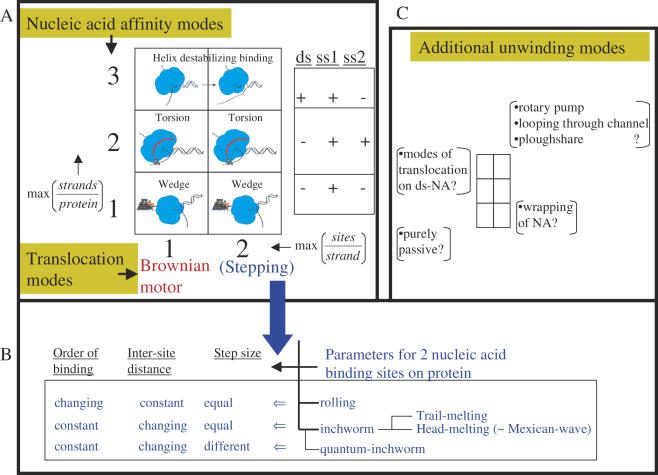
Figure 4.
Possible mechanisms of helicase translocation and nucleic acid unwinding. Nucleic acid unwinding is considered to be a combination of two processes: unidirectional translocation (achieved by the protein via polar binding to DNA) and strand separation (a function of transient sets of nucleic acid affinity modes of the protein). The six-part box in (A) illustrates the possible unwinding modes resulting from the various nucleic acid binding modes (y-axis) and translocation modes (x-axis). The second box on the right in (A) shows an experimental scheme that parallels the three nucleic acid binding modes of the y-axis. For example, if the only type of binding between the protein and DNA is one protein site bound to one strand of the DNA, then the stoichiometry max (strands/protein) would be 1. If the maximal stoichiometry is two strands per protein the value would be 2, and if one protein can bind to both duplex and single stranded nucleic acid, the y-axis value would be 3 (= 2 + 1). The nucleic acid affinity modes outlined on the y-axis define the unwinding mechanisms depicted with pictures within the boxes. The wedge unwinding mode results from the helicase interacting exclusively with one strand, a torsional component of unwinding would necessitate interaction with an additional strand of nucleic acid, and direct destabilization of the nucleic acid junction would require interaction with the duplex region at the junction in addition to the single strand on which the helicase is translocating. The x-axis defines the translocation mechanisms, and the two numerical values it can take correspond to the maximum observable number of nucleic acid sites that the helicase can transiently and simultaneously bind to during translocation. If there is only one possible nucleic acid binding site, the helicase may move unidirectionally along the nucleic acid using a Brownian motor (BM) or ratchet type mechanism (68,69). If the helicase binds the nucleic acid via two sites, it can employ stepping modes of translocation, such as rolling or inchworm. The stepping translocation modes are further elaborated in (B). In the rolling mechanism, the two helicase sites that bind successive regions of the nucleic acid alternate their affinity while retaining the distance between them but not their relative order. In other words, one helicase site is ahead in one step, but the other one moves in front during the next step. In the inchworm mechanism, the helicase polarity is maintained during translocation; thus, the same side of the protein faces the same end (3′ or 5′) of the nucleic acid at all times. To move, the distance between the two protein sites and the binding affinity periodically changes. A widely accepted mechanism combines the inchworm translocation mode with the duplex destabilization mode. In this mechanism, the two contact sites, protein–dsDNA and protein–ssDNA, can be in one of two relative orders. If the dsDNA binding site is ahead of the other, the mechanism might be referred to as head-melting, or trail-melting in the other case. The mode of head-melting can be likened to a Mexican-wave movement along the DNA (70,71). In the case where these two sites take steps of different sizes, the mechanism was given the name quantum inchworm (72). Further modifications of inchworm mechanism taking into account requirements for the nature of the displaced strand or the predominant mode of interaction with the loading strand (i.e. hydrophobic or electrostatic) have also been proposed (73–75). We display all six combinations as physically plausible, even though not all have been proposed previously. The most popular mechanisms are shown by the coordinates (1,1), (1,2) and (2,3).In addition to the above modes, several other types of unwinding mechanisms have been proposed as illustrated in (C). For example a purely passive mechanism is probably employed by the chemical formaldehyde (76), ssRNA wrapping might be used by Rho, dsDNA translocation might be used by DnaB during branch migration (15), ploughshare is proposed for MCM proteins (77), and looping through channel is proposed for SV40-large T antigen (78). In addition, the torsional model can work with a duplex DNA in the central channel in addition to the mentioned mode of binding two single strands separately. Classifying most of these specific mechanisms requires extra dimensions in the 2 × 3 graph of part (A).