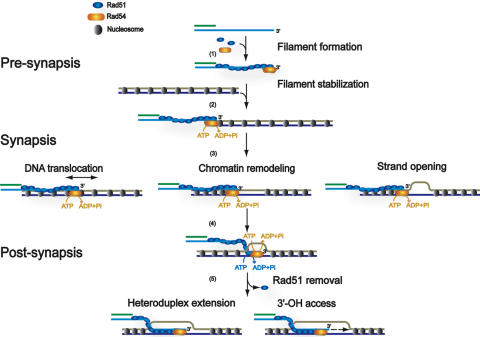
Figure 3.
Mechanistic models for Rad54 function in HR. The mechanistic models were derived from analysis of reconstituted in vitro recombination reactions and biochemical analysis of the Rad54 protein. For more details see text. Shown is one processed DSB end with a 3′-ending ssDNA tail that invades a nucleosomal duplex target DNA. Pre-synapsis: Rad54 was found to mediate formation or to stabilize Rad51 filaments on ssDNA. The pre-synaptic function does not require Rad54 ATPase activity and requires Rad51 binding to ATP but not hydrolysis. Synapsis: Rad54 augments the ability of Rad51-ssDNA filaments to form joint molecules, possibly involving translocating the Rad51-ssDNA filament along duplex DNA or inducing strand separation through induction of topological change. Rad54 also exhibits chromatin remodeling activity that may clear nucleosomes or other proteins from the pairing site. The synaptic function requires the Rad54 ATPase activity but not the Rad51 ATPase activity. Post-synapsis: Rad54 was identified to catalyze heteroduplex extension (branch migration) and can dissociate the Rad51–dsDNA product complex, possibly to allow DNA polymerase access to the invading 3′-OH end to prime DNA synthesis. Post-synapsis requires the ATPase activities of both the Rad54 and Rad51 proteins. The exact oligomeric structure of Rad54 in its interaction with the Rad51-ssDNA and Rad51–dsDNA filaments is not known, so it is unclear how many molecules of Rad54 are represented by the symbol drawn in the figure. Rad54 is likely to act as an oligomer on DNA (58), and an oligomeric Rad54 particle has been directly visualized at the terminus of Rad51–dsDNA filaments by electron microscopy (81). This figure is derived from Figure 2 of reference (12).