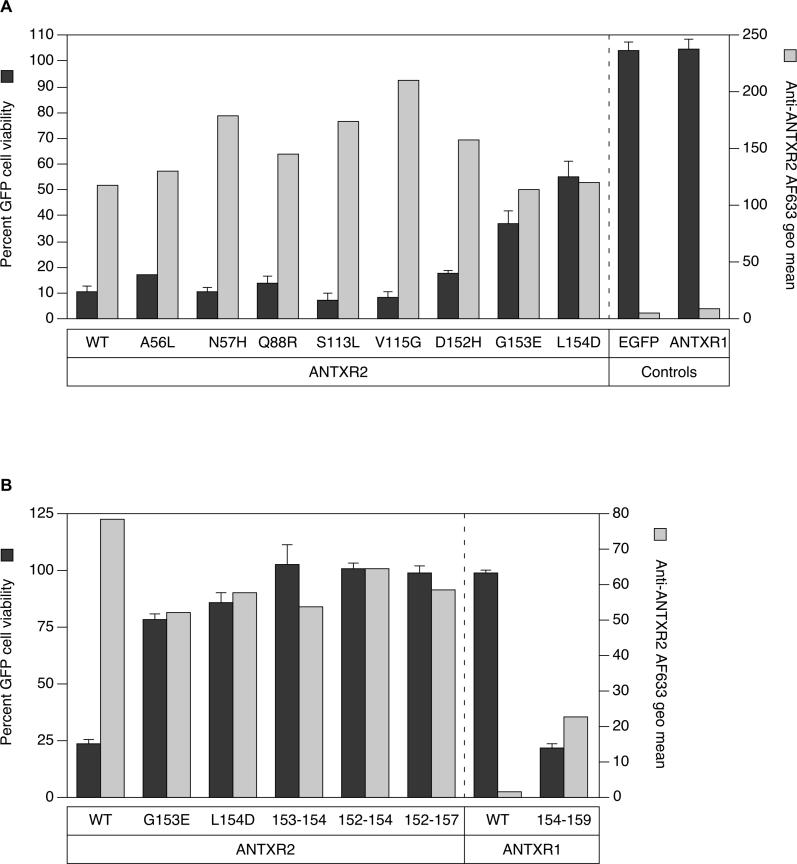
Figure 3. ANTXR2 Contact with PA Domain 2 Confers Sensitivity to PAD683N-Mediated Intoxication.
(A) Triplicate samples of CHO-R1.1 cells transiently expressing ANTXR2, or ANTXR2 proteins with alterations in PA contacts to nonconserved ANTXR1 residues were incubated with 10−10 M LFN-DTA and 10−7 M PAD683N, or LFN-DTA only, and analyzed by flow cytometry. The average percentage of live EGFP-positive cells in the samples with both parts toxin is divided by that in the LFN-DTA only samples to determine percent cell viability (black bars). Individual samples of the same cells were also incubated with an anti-ANTXR2 antibody, followed by an AlexaFluor-633–conjugated secondary antibody and analyzed by FACS to determine the receptor cell surface expression level. The geometric mean of AlexaFluor-633 fluorescence after staining cells with antibodies (gray bars) is depicted next to the cell viability data for comparison purposes.
(B) Receptor sequences responsible for contacting PA domain 2 were exchanged between ANTXR1 and ANTXR2 proteins in an analysis similar to (A). Cells expressing ANTXR1154–159 bound anti-ANTXR2 antibodies, suggesting that an epitope for the polyclonal antibody mapped to this site. Saturable binding studies performed with transfected cells and with increasing amounts of an AlexaFluor-633–labeled monomeric PA protein confirmed that the mutant 154–159 ANTXR1 protein is expressed on the cell surface at levels similar to wild-type ANTXR1 and ANTXR2 (unpublished data).