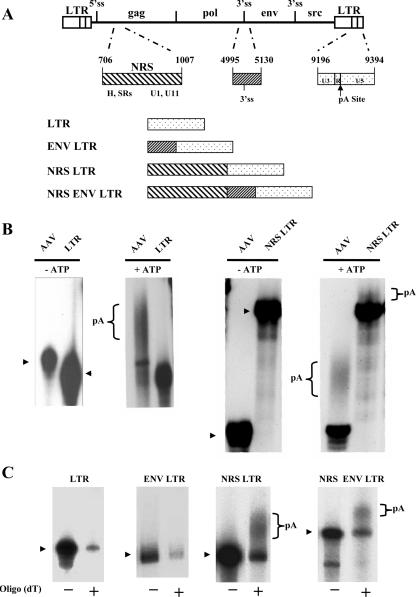
FIG. 1.
The NRS promotes polyadenylation in vitro. (A) Schematic representation of the RSV proviral genome with an expanded view of the NRS, the region flanking the env 3′ splice site, and the portion of the LTR used in this study. The U3 region of the LTR begins at nucleotide 9058, and the major cleavage/polyadenylation (pA) site is at nt 9312 (arrow). hnRNP H and SR proteins have been shown to bind the 5′ end of the NRS, whereas U1 and U11 snRNPs bind at the 3′ end. Polyadenylation substrates are schematically represented. (B) In vitro polyadenylation assays. The indicated radiolabeled, capped RNAs were incubated in in vitro polyadenylation reaction mixtures containing HeLa cell nuclear extract in the presence or absence of ATP. The arrowheads indicate the precursor RNA. (C) After in vitro polyadenylation reactions were performed with the indicated transcripts, poly(A)+ RNA was isolated using an oligo(dT) column. −, unpolyadenylated RNA that flowed through the column; +, poly(A)+ RNA eluted. The arrowhead at the left of each autoradiogram indicates the precursor RNA. Polyadenylated transcripts in the NRS LTR and NRS ENV LTR are denoted reactions.