Figure 1.
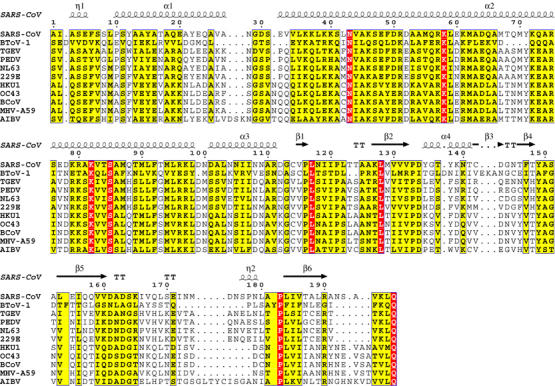
Sequence alignment of nsp8 proteins. The alignment of coronavirus nsp8 sequences was generated with the ClustalW program, version 1.82 (http://www.ebi.ac.uk/clustalw/). This alignment and individual nsp8 sequences were used to search sequence databases as described in Snijder et al (2003). Using results of these searches, the original alignment was extended to a distantly related (see text) torovirus sequence using the MUSCLE program. The resulting alignment was converted into this figure using the ESPript program, version 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Residues that are conserved in all or >70% sequences are boxed in red and yellow, respectively. Above the alignment, numbering and secondary structure elements (Zhai et al, 2005) for SARS-CoV nsp8 protein are depicted. National Center for Biotechnology Information (NCBI) Accession Numbers for replicase polyprotein sequences including nsp8: SARS-CoV, NP_828866; Breda torovirus (BToV-1), AY427798; Transmissible Gastroenteritis coronavirus (TGEV), NP_840006; Porcine epidemic diarrhea virus CV777 (PEDV), NP_839962; Human coronavirus NL63 (NL63), AAS58176; Human coronavirus 229E (229E), NP_835349; Human coronavirus HKU1 genotype A (HKU1), AAT98578; Human coronavirus OC43 (OC43), NP_937947; Bovine coronavirus (BCoV), NP_742135; Mouse Hepatitis virus strain A59 (MHV-A59), NP_740613; Avian Infectious Bronchitis Virus (AIBV), NP_740626.
