Figure 7.
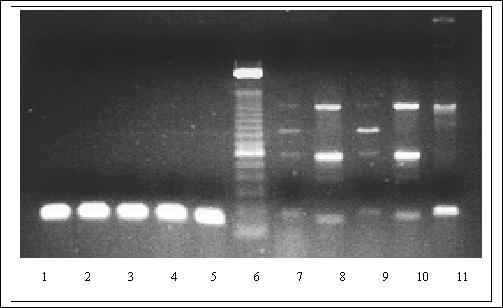
Demonstration of specificity of designed PCR primers for Pseudomonas. The following oligonucleotides were designed as forward 5'-CTACGGGAGGCAGCAGTGG-3'and reverse 5'-TCGGTAACGTCAAAACAGCAAAGT-3' for PCR. Lane 1–5: Five different reported Pseudomonas strains: SF1 & PH1 (laboratory isolates), Pp2 & CF600 (Received from Dr. V. Shingler, Umea university, Sweden), P. putida (received from Dr. S. Harayama, MBI, Japan); Lane 6: 100 bp ladder; Lane 7: E. coli, Lane 8: Salmonella, Lane 9: Vibrio, Lane 10: Shigella Lane 11: Staphylococcus. A PCR program of 95°C, 5 min, and 35 cycles of 94°C, 30 s; 62°C, 15 s; and 72°C for 30 s were used to amplify a 150 bp product using total DNA as template. The designed primers were used at 25 pM concentration of each in 50 μl reaction mixture as per the conditions described by the supplier (Perkin Elmer). Lane 1–5 shows the amplification of expected Pseudomonas specific product; and lanes 7–11 the non-specific extension of primers with strains negative for the selected conserved region. The quality of the DNA as a template and the control for PCR were checked using the universal eubacterial primers (data not shown).
