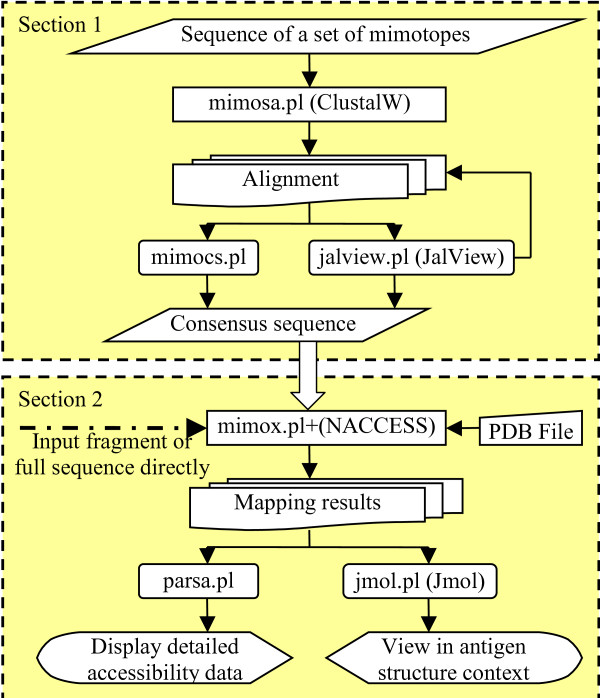
Figure 1.
Overall architecture of MIMOX. MIMOX has two sections. The first section has 3 perl scripts. The script mimosa.pl aligns a set of mimotope sequences powered by ClustalW. The script jalviews.pl wraps JalView to view and manage the alignment. The script mimocs.pl derives a consensus sequence from the alignment. The second section also has 3 perl scripts. The script mimox.pl maps the user supplied sequence on to the given antigen structure and utilizes NACCESS to calculate the accessibility. The script parsa.pl displays the detailed accessibility information of each mapping result. The script jmol.pl wraps Jmol to view the mapping result interactively on the antigen structure.