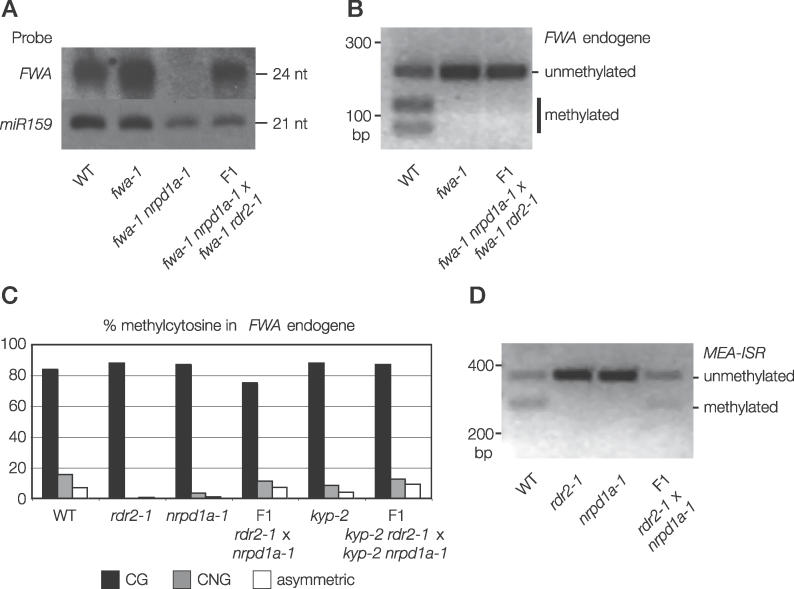
Figure 5. Separable Recruitment of siRNA Production and Non-CG DNA Methylation at Chromosomal Tandem Repeats.
(A) Unmethylated chromosomal tandem repeats can recruit siRNA production. fwa-1 nrpd1a-1 and fwa-1 rdr2–1 lack FWA siRNAs, but when these mutants are crossed together, Northern blotting shows that FWA siRNA production is restored in F1 plants.
(B) Unmethylated chromosomal tandem repeats cannot recruit de novo DNA methylation. When fwa-1 nrpd1a-1 and fwa-1 rdr2–1 mutants are crossed together, FWA remains unmethylated despite resumption of siRNA production. DNA methylation of FWA was assayed by PCR from bisulfite-treated DNA followed by ClaI digestion.
(C) CG DNA methylation recruits siRNA-directed non-CG DNA methylation to FWA. When the recessive rdr2–1 and nrpd1a-1 mutants are crossed together, non-CG DNA methylation returns to CG-methylated FWA in the F1 plants. DNA methylation was assayed by bisulfite sequencing.
(D) CG DNA methylation recruits siRNA-directed non-CG DNA methylation to MEA-ISR. Non-CG DNA methylation returns to CG-methylated MEA-ISR in rdr2–1 × nrpd1a-1 F1 plants. Asymmetric DNA methylation was assayed by PCR from bisulfite-treated DNA followed by BamHI digestion. DNA methylation protects the BamHI site from bisulfite conversion, allowing restriction digestion after bisulfite treatment.