Fig. 4.
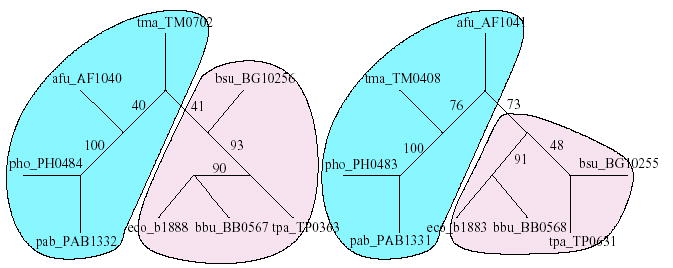
Evolutionary trees (not drawn to scale) of cheA–bacteria and cheB–bacteria obtained from multiple sequence alignment using CLUSTALW v1.83. The numbers along the internal edges represent the bootstrap values of the edges. The tree topologies are clearly different, defying the hypothesis that the evolutionary trees of families (that are known to interact) have similar topologies. Even though the corresponding left subtrees are topologically identical, the positioning of ‘tma_TMxxxx’ and ‘afu_Afxxxx’ proteins is not the same.
