Figure 1.
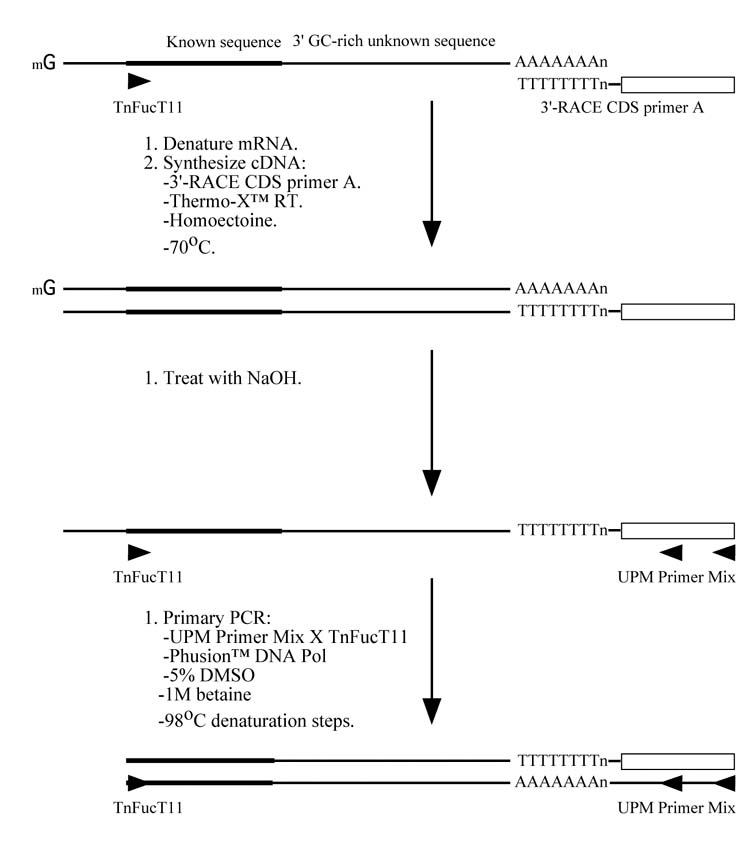
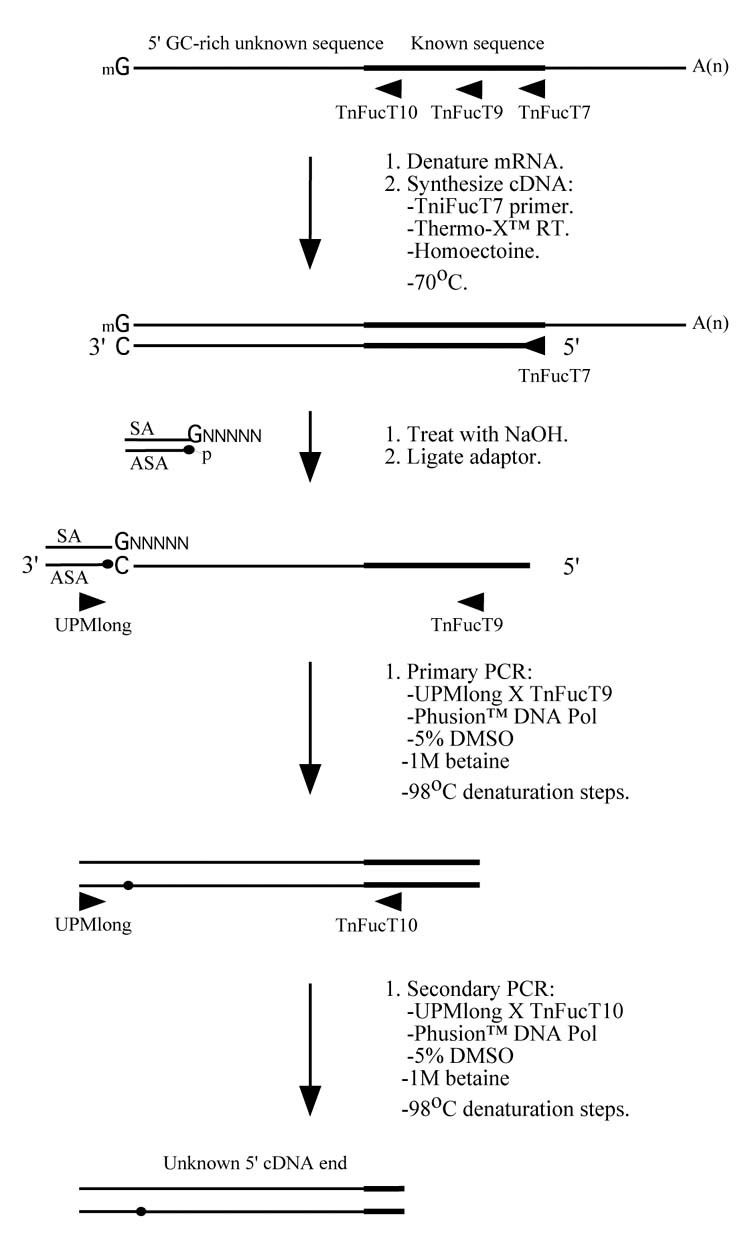
A new RACE method for extremely GC-rich genes. (A) For 5′-RACE, the first strand cDNA is synthesized at 70°C using Thermo-X reverse transcriptase in the presence of 0.4875 M homoectoine and a gene specific primer (in this case, FucT7). The mRNA is then hydrolyzed and the cDNA is denatured in preparation for the addition of an adaptor, which provides an anchor sequence that can be used for subsequent semi-nested PCRs. The primary and secondary PCRs are performed using nested internal gene specific primers (in this case, TniFucT9 and TniFucT10), a primer complementary to the antisense adaptor sequence, ASA (in this case, a commercial primer called UPMlong), Phusion™ DNA polymerase, and a denaturation temperature of 98°C. (B) For 3′-RACE, the first strand cDNA is synthesized at 70°C using Thermo-X reverse transcriptase in the presence of 0.4875 M homoectoine and a primer complementary to the poly-A tail of the mRNA (in this case, a commercial primer called 3′-RACE CDS primer A). The mRNA is then hydrolyzed and a PCR is performed using a gene specific primer (in this case, TnFucT11) and a primer complementary to the specific portion of the oligonucleotide used to prime the reverse transcription reaction (in this case, a commercial primer called UPM primer mix). The other conditions used for the PCR are identical to those used for 5′-RACE and nested PCR may be used, if required.
