Abstract
The multifunctional protein NS1 of minute virus of mice (MVMp) is posttranslationally modified and at least in part regulated by phosphorylation. The atypical lambda isoform of protein kinase C (PKCλ) phosphorylates residues T435 and S473 in vitro and in vivo, leading directly to an activation of NS1 helicase function, but it is insufficient to activate NS1 for rolling circle replication. The present study identifies an additional cellular protein kinase phosphorylating and regulating NS1 activities. We show in vitro that the recombinant novel PKCη phosphorylates NS1 and in consequence is able to activate the viral polypeptide in concert with PKCλ for rolling circle replication. Moreover, this role of PKCη was confirmed in vivo. We thereby created stably transfected A9 mouse fibroblasts, a typical MVMp-permissive host cell line with Flag-tagged constitutively active or inactive PKCη mutants, in order to alter the activity of the NS1 regulating kinase. Indeed, tryptic phosphopeptide analyses of metabolically 32P-labeled NS1 expressed in the presence of a dominant-negative mutant, PKCηDN, showed a lack of distinct NS1 phosphorylation events. This correlates with impaired synthesis of viral DNA replication intermediates, as detected by Southern blotting at the level of the whole cell population and by BrdU incorporation at the single-cell level. Remarkably, MVM infection triggers an accumulation of endogenous PKCη in the nuclear periphery, suggesting that besides being a target for PKCη, parvovirus infections may also affect the regulation of this NS1 regulating kinase. Altogether, our results underline the tight interconnection between PKC-mediated signaling and the parvoviral life cycle.
The regulation not only of cellular proteins but also of viral proteins by phosphorylation has attracted research interest for many years. Besides characterization of proteins which become phosphorylated and the identification of their regulatory kinases, much effort has been spent on the analysis of the signaling pathways involved and their functional consequences. We are particularly interested in understanding how the multifunctional nonstructural protein NS1 of the autonomous parvovirus minute virus of mice (MVMp) is regulated. MVM consists of a small icosahedral capsid with a linear single-stranded DNA of negative polarity as a genome. The DNA of MVMp codes for the nonstructural proteins NS1 and NS2, of which the latter exists in three different isoforms, differing in their unique C termini only, as well as two capsid proteins, VP1 and VP2. NS1 is endowed with numerous biochemical activities, such as ATP binding and hydrolysis (12, 62), helicase (44, 62), site-specific binding to the cognate recognition motif [ACCA]2-3 which is scattered throughout the viral genome (13, 19), and site- and strand-specific endonuclease (11, 18, 44). Furthermore, NS1 takes part in protein-protein interactions to form homo-oligomers (40, 52) or complexes with cellular partner proteins like the transcription factor SP1 (35), the co-chaperone SGT (20, 58), or heterogeneous nuclear ribonucleoproteins (28). NS1 plays a key role in many processes necessary for progeny virus production. Thus, it initiates and regulates viral DNA amplification (for a review, see reference 17) or trans regulates its own P4 promoter (22) as well as the P38 promoter driving capsid gene expression (13, 53). In addition, NS1 exerts cytotoxic stress on its host cells, which shows itself in cell cycle arrest (49), disregulation of heterologous promoters (36, 60), and changes in cell physiology (1) and morphology (9, 14). Such a diversity of activities implies a tight regulation of NS1 functioning. Indeed, phosphorylation proved to serve as a mechanism for coordination of the various NS1 functions in a time-dependent (15) and subcellular location-dependent (46) manner. Thus, modulations of the NS1 phosphorylation state altered its biochemical profile (43), and NS1 mutagenesis at consensus (protein kinase C [PKC]) phosphorylation sites selectively impaired the viral polypeptide in some of its functions necessary for virus propagation (14, 21). Although NS1 comprises more than 100 potential consensus phosphorylation sites and generates more than 20 distinct phosphopeptides when isolated from infected cells (15), only a few phosphorylated residues of NS1 (14, 21) and a single cellular kinase, namely the atypical PKCλ (21, 46), could be identified so far as being involved in the regulation of the viral protein.
The molecular mechanisms of parvoviral DNA replication have been extensively investigated (for a review, see reference 17). Replication of the single-stranded genome occurs through double-stranded concatemeric DNA intermediates as a result of a rolling circle mode of replication (RCR)-like mechanism similar to the one described for bacteriophages, single-stranded plasmids, and geminiviruses (for a review, see reference 34). Upon entering the nucleus, virion DNA becomes converted into a covalently closed monomeric duplex, which serves as a first transcription template for production of viral proteins, including NS1. In concert with cellular accessory proteins, NS1 generates a free 3′ hydroxyl group through its site- and strand-specific nicking activity. Thereafter, polymerase δ drives the formation of concatemeric duplex intermediates by a unidirectional, leading-strand elongation mechanism. To facilitate strand-displacement synthesis, NS1 works as a processive helicase, unwinding the double-stranded replication template in front of the replication fork. For both the initial nicking reactions at the left- and right-end origins and viral DNA amplification, NS1 physically interacts with cellular components, such as the transcription factor PIF (11), members of the HMG family (18), and components of the replication machinery, such as RPA and RF-C (10).
The regulation of NS1 enzymatic functions in particular during replication of the viral genome has been characterized in detail by using both site-directed mutagenesis (21, 42) and functional in vitro assays in which dephosphorylated inactive NS1 was rescued through incubation with consecutively purified cellular extracts or recombinant protein kinases (43, 45, 46). Phosphorylation of NS1 residues T435 and S473 by PKCλ regulates the site-specific binding and DNA unwinding activities of the viral polypeptide. These phosphorylation events also determine the ability of NS1 to site-specifically nick the left-end MVM origin (42) and control strand-displacement synthesis through its processive helicase function (21). Yet to become fully competent for RCR activity, NS1 requires additional phosphorylation steps executed by an as-yet-undefined TPA-responsive cellular protein kinase (21, 45).
In this paper we aimed to identify additional kinases that are required for complete activation of NS1 RCR functions. Therefore, the strategy previously described for identification of atypical PKCλ (45) was applied. Distinct fractions of cellular extracts were tested for their ability to rescue dephosphorylated NS1 in a kinase-free replication system and analyzed for the presence of candidate protein kinases. Finally, fractionation on hydroxyl apatite columns led to the identification of the novel PKCη as a prime candidate for the regulation of NS1 RCR functions. The competence for NS1 activation was then confirmed through cloning of the PKCη cDNA into a mammalian expression system and testing of the purified recombinant protein in a kinase-free in vitro replication system. Furthermore, the impact of phosphorylation of NS1 by PKCη on MVM DNA replication was verified in vivo, using stably transfected cell lines endowed with an altered PKCη activity upon infection. Altogether, our results provide strong evidence for the involvement of an additional member of the PKC family, novel PKCη, in the regulation of NS1 for viral DNA amplification in cooperation with atypical PKCλ.
MATERIALS AND METHODS
Antibodies and reagents.
Polyclonal antibodies against PKCβI and PKCβII, PKCδ, PKCɛ, PKCη, or PKCζ (C-16, C-18, C-17+C-20, E-5, C-15, and N-17) were purchased from Santa Cruz. Monoclonal antibodies recognizing PKCα, PKCγ, PKCλ, or PKCμ were obtained from Transduction Laboratories. The PDK-1-specific sheep polyclonal antibody (06-906) was from Upstate Biotechnology. Horseradish peroxidase-conjugated antirabbit and antimouse antibodies were purchased from Promega, while horseradish peroxidase-conjugated antisheep antibody was obtained from Upstate Biotechnology. Fluorescent dye-labeled secondary antibodies were from Dianova, with the exception of the Cy3-conjugated antirat-antibody purchased from MoBiTec. Rat antibromodeoxyuridine (anti-BrdU) antibodies were obtained from Direct.com.
Cells and viruses.
BSC-40, A9 cells, and derivatives thereof were maintained as monolayers in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum. HeLa-S3 cells were grown in spinner bottles in the presence of 10% fetal calf serum. MVMp was propagated in adherent A9 cells, and virus stocks were prepared by repeated freezing and thawing in 10 mM Tris (pH 8.3)-1 mM EDTA. Recombinant vaccinia viruses were constructed and propagated as previously described (41, 43). Vaccinia viruses expressing His-tagged NS1 (44) and human (h) or mouse (m) PKCs hPKCα, hPKCγ, mPKCλ, and hPKCζ (21) have been described earlier.
Cloning of PKC cDNAs.
Human placenta and A9 cDNA libraries were generated from mRNA preparations using a SMART PCR cDNA synthesis kit (Becton Dickinson, Heidelberg, Germany). PKCη was cloned by PCR from the human placenta library in two separate fragments. The N-terminal fragment was obtained using the primer pair A (5′-ATGTCGTCTGGCACCATGAAGTTCAATGGCTATTTGAGGGTCCG-3′) and B (5′-CCACAGTTAGGGGCCACGTTCGCTTGACATCGAATATGCA-3′); for the C-terminal half of the coding region, primers C (5′-TGCATATTCGATGTCAAGCGAACGTGGCCCCTAACTGTGG-3′) and D (5′-CTATGGTTGCAATTCTGGAGACACATAGGAAAAGTTTCTA-3′) were used. The two fragments were then combined through the overlapping sequences of primers B and C by additional PCR and cloned directly into pCR2.1 (Invitrogen). Similarly, mouse PKCδ and mouse PKCɛ cDNAs were amplified from the A9 cDNA library. For PKCδ, the following primers were used: A (5′-ATGGCACCCTTCCTGCGCATCTCCTTCAATTCCTATGAGC-3′), B (5′-CCCGGCATTTGTGGTGCACATTCATGCCACAATCTTCACA-3′), C (5′-TGTGAAGATTGTGGCATGAATGTGCACCACAAATGCCGGG-3′), and D (5′-TTAAATGTCCAGGAATTGCTCAAACTTGGGATTCACAAAG-3′). For PKCɛ we used primers A (5′-ATGGTAGTGTTCAATGGCCTTCTTAAGATCAAAATCTGCG-3′), B (5′-CTCCGGGGCTTGCCAGCTGGCCATCGGTGGCCGACGACGC-3′), C (5′-GCGTCGTCGGCCACCGATGGCCAGCTGGCAAGCCCCGGAG-3′), and D (5′-TCAGGGCATCAGGTCTTCACCAAAGTAGGAGAAGCCTTTA-3′). All pCR2.1-cDNA clones were sequenced; no differences within the coding sequences were found compared to the GenBank sequences (hPKCη, NM_006255; mPKCδ, AB011812; and mPKCɛ, AF028009).
Plasmid constructs for recombinant PKCη expression. (i) Production of purified proteins (see below).
Plasmid pTMHis, a derivative of pTM-1 which allows the expression of N-terminally His6-tagged proteins (45), was used as an expression vector. To generate the pTHisPKCx plasmids, PCRs were carried out using the above-mentioned PKC clones together with N- and C-terminal primers harboring unique restriction sites compatible with the polylinker in pTMHis1. The N-terminal restriction site was designed to fuse the His6 tag in frame with the coding sequence of the PKC cDNA. All PCRs were performed according to the manufacturer's recommended conditions using the Advantage HF PCR amplification kit (Becton Dickinson), and products were first subcloned into pCR2.1 (Invitrogen) prior to transfer into the expression vector. The hPKCη-coding PCR fragment was generated with the primers 5′-TACGGATATCCATGGCGTCTGGCACCATGAAGTTCAATG-3′ and 5′-TACGTCTAGATATCCTATGGTTGCAATTCTGGAGACACA-3′ using pCR2.1-hPKCη as a template. This fragment was then cloned as an NcoI-to-EcoRV segment into NcoI- and SmaI-cleaved dephosphorylated pTHis1, giving rise to pTHis hPKCη.
The mPKCδ coding fragment (primers 5′-GATATCGAATTCATGGCACCCTTCCTGCGCATCTCCTTCA-3′ and 5′-TCTAGATTAAATGTCCAGGAATTGCTCAAA-3′) was transferred as an EcoRI segment into EcoRI-cleaved, dephosphorylated pTHis1, giving rise to pTHismPKCδ. The mPKCɛ sequence was transferred as an NcoI-to-XhoI PCR fragment (primers 5′-TCAGCCATGGTAGTGTTCAATGGCCTTCTTAAGATC-3′ and 5′-TCAGCTCGAGTCAGGGCATCAGGTCTTCACCAAAGT-3′) into similarly digested pTHis1, generating pTHismPKCɛ.
For PKC expression in stably transfected cells, we used plasmid pP38, obtained by deleting the PmeI-to-PvuII fragment from pdbMVMpdl1200 (33). This construct drives the expression of foreign genes under control of the parvoviral NS1 protein. For generation of N-terminally Flag-tagged fusion proteins, we used plasmid pP38-Flag, a pP38 derivative that was obtained by inserting the polylinker (5′-GCTAATGGCTGACTACAAGGACGACGATGACAAGGCCAAGCTTCGAATTCTGCAGTCGACGGTACCGCGGGCCCGGGAT(X)nGCGGCCGC-3′) as an Eco47III and NotI fragment into HpaI- and NotI-digested pP38. The polylinker comprises the Flag epitope (MADYKDDDDKA) and allows the in-frame cloning of foreign sequences through restriction sites for HindIII, EcoRI, SalI, AccI, KpnI, SacII, ApaI, and SmaI. The mutant PKCη-cDNA clones (see below) were subcloned into pCR2.1 (Invitrogen, Groningen, The Netherlands) prior to transfer into the expression vector pP38-Flag as SmaI-NotI (PKCηA160E and T512A) fragments or as a SmaI-to-XbaI (PKCη-DN) fragment, respectively. To generate the control pP38GFP plasmid, the coding sequence of enhanced green fluorescent protein (EGFP) (Clontech) was transferred as a StuI-to-XbaI-cleaved PCR fragment (primers 5′-AGCTAGGCCTCCATGGTGAGCAAGGGCGAGGAGCTGTTC-3′ and 5′-ATCGCCCGGGTCTAGAGTCGCGGGCCGCTTTAC-3′) into HpaI- and XbaI-cleaved pP38.
(ii) Production of mutant forms of PKCη.
Site-directed mutagenesis of the hPKCη cDNA clone was performed by chimeric PCR using the N- and C-terminal primers 5′-CGG GCC CGG GAT ATG TCG TCT GGC ACC ATG AAG-3′ and 5′-GGCGCGCGGCCGCCTATGGTTGCAATTCTGGAGAC-3′, together with two overlapping internal primers harboring the mutation. These mutated primers were the following: for PKCηA160E (replacing alanine with glutamic acid in the pseudosubstrate region), 5′-CAG GAA GCG CCA AAG GGA AAT GCG AAG GCG AGT CCA CC-3′ and 5′-GGA CTC GCC TTC GCA TTT CCC TTT GGC GCT TCC TGG TA-3′; and for PKCηT512A (replacing the PDK phosphorylation site threonine with alanine), 5′-GTC ACC ACG GCC GCA TTC TGT GGC ACG CCA GAC-3′ and 5′-GGC GTG CCA CAG AAT GCG GCC GTG GTG ACA CCA TTG-3′ (boldface shows mutated residue). Another mutant, PKCη-DN, encoding only the regulatory domain of PKCη (amino acids [aa] 1 to 296) was generated in a single PCR run using the primer pair 5′-CGG GCC CGG GAT ATG TCG TCT GGC ACC ATG AAG-3′ and 5′-TACGTCTAGATATCTTACCCCACAGTTAGGGGCCACGTT-3′. Due to primer-related reasons, the C terminus of PKCηDN was extended by the following amino acids: RYLELLHMVHFSGKEN. Before transferring into appropriate expression vectors, all three PKC mutants were verified by sequencing.
Protein extraction and fractionation by column chromatography.
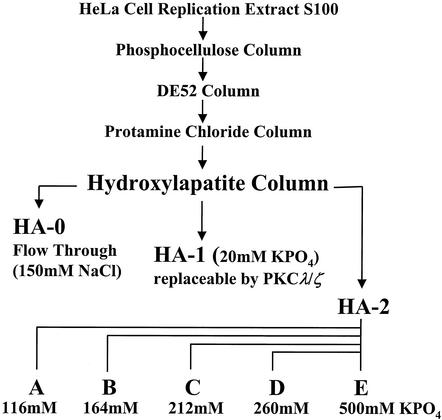
S100 extracts from HeLa cells were prepared and successively fractionated on phosphocellulose, DE52, and protamine (PA) chloride columns as previously described (45). The PKC-containing fraction PA-2 was eluted from the column using buffer C (20 mM HEPES [pH 7.5], 1 mM EDTA, 0.1 mM dithiothreitol [DTT], 10% glycerol) containing 1 M NaCl and the protease inhibitors phenylmethylsulfonyl fluoride (174 μg/ml), leupeptin (5 μg/ml), and aprotinin (2 μg/ml). PA-2 was dialyzed, adjusted to 50% glycerol, and stored in aliquots at −80°C. PKC isoforms present in fraction PA-2 were separated by fast-performance liquid chromatography (Pharmacia) on hydroxyl apatite (HA) columns (Merck). PA-2 (corresponding to a 3-liter culture of ∼2 × 109 cells) was adjusted to 200 mM NaCl, loaded on a 5-ml HA column with constant flux (0.5 ml/min), and washed with 30 ml of buffer C containing 50 mM NaCl. After collection of the flowthrough (HA-0) and wash, the HA-1 fraction (containing the atypical PKC isoforms PKCλ and PKCζ) was eluted from the column using buffer D (150 mM NaCl, 20 mM KPO4 [pH 7.5], 10% glycerol, protease inhibitors phenylmethylsulfonyl fluoride, leupeptin, aprotinin). The bound material (HA-2) comprising the nondefined component(s) necessary for NS1 activation in RCR assays was then further fractionated using a step-gradient of KPO4 as indicated in Fig. 1. All fractions were dialyzed against buffer C containing 50 mM NaCl overnight at 4°C, adjusted to 50% glycerol, and frozen in aliquots at −80°C.
FIG. 1.
Purification of NS1-regulating kinases from crude HeLa cell extracts. Protein kinases regulating NS1 for replicative functions were purified on phosphocellulose, DE52, protamine chloride, and HA columns as described previously (45). To identify the NS1O-activating protein kinases, the protamine-bound components were fractionated by HA chromatography, using a step gradient, increasing KPO4 concentrations as indicated. Atypical PKCλ/ζ, necessary to activate NS1O for DNA unwinding activities, eluted at 20 mM KPO4 (HA-1 fraction). Besides HA-1, the HA-2 fraction was required to fully activate NS1 for RCR. To further separate protein kinases with higher affinity to HA (HA-2 fraction), elution was carried out at higher KPO4 concentrations, yielding subfractions A to E.
Production and purification of recombinant proteins by means of vaccinia virus expression.
NS1 and recombinant PKC isoforms were produced by means of vaccinia virus expression in suspension cultures of HeLa-S3 cells and harvested around 18 h postinfection. His-tagged NS1 present in nuclear extracts was dephosphorylated, or not, with calf intestine alkaline phosphatase and purified immediately on Ni2+-nitrilotriacetic acid agarose columns (43). The protein preparations were analyzed by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected either by Coomassie blue staining (staining buffer: 25% methanol, 10% acetic acid, 0.1% Coomassie brilliant blue; destaining buffer: 25% methanol, 10% acetic acid) or by Western blotting.
In vitro replication assays.
Template plasmids pL1-2TC and pL1-2GAA containing the minimal active left-end MVM origin and the corresponding inactive origin, respectively, were described and characterized previously (16).
Replication assays were carried out as described previously (45) in the presence of optimized P1-Thr subcellular fractions, 3 U of T4 DNA polymerase, and approximately 200 ng of His-tagged vaccinia virus-produced NS1 (as determined by Coomassie blue staining after SDS-PAGE). P1-Thr consists of the flowthrough fraction of 293 cell extracts purified on phosphocellulose columns and relieved of endogenous serine-threonine kinases by l-Thr-affinity chromatography. This fraction contains the replication factors RPA, PCNA, and PIF. The assays were carried out in a 20-μl total volume of 20 mM HEPES-KOH (pH 7.5), 5 mM MgCl2, 5 mM KCl, 1 mM DTT, 0.05 mM (each) deoxynucleoside triphosphates; 2 mM ATP, 40 mM creatine phosphate, 1 μg of phosphocreatine kinase, 10 μCi of [α-32P]dATP (3,000 mCi/mmol), and 20 ng of the appropriate DNA template (pL1-2TC or pL1-2GAA). After incubation at 37°C for 2 h, the reaction was stopped by adding 60 μl of 20 mM Tris (pH 7.5)-10 mM EDTA-0.2% SDS and heating the mixture to 70°C for at least 30 min. The reaction products were analyzed by agarose gel electrophoresis after immunoprecipitation with anti-NSN antiserum and digestion with HindIII.
In vitro kinase reactions.
In vitro kinase reactions were performed as described previously (43). Dephosphorylated NS1O (200 ng) was incubated with purified recombinant PKCζ or PKCη (100 ng) in the presence of 20 mM HEPES-KOH (pH 7.5), 7 mM MgCl2, 5 mM KCl, 1 mM DTT, 10 μCi of [γ-32P]ATP (3 Ci/mmol) and PKC cofactors (1 μg of l-α-phosphatidyl-l-serine/ml, 1 nM TPA). After incubation for 30 min at 37°C, the reactions were stopped by adding the same volume of 20 mM Tris (pH 7.5)-5 mM EDTA-0.2% SDS, and heating for 30 min at 70°C. The reaction products were immunoprecipitated with anti-NSN antiserum and analyzed by SDS-10% PAGE and autoradiography after blotting on nitrocellulose or polyvinylidene difluoride membranes.
Generation of stably transfected A9 cell lines.
Stable transfectants were generated by cotransfection of 105 A9 cells with 25 μg of the appropriate pP38-X construct together with pSV2neo, in a molar ratio of 25:1, using 25 μl of Lipofectamine (Invitrogen) according to the manufacturer's protocol. Two days posttransfection, cultures were split 1:10, and transfected cells were selected using 400 μg of G418 (Sigma)/ml. Colonies were pooled after growth for approximately 4 weeks under selection, and frozen stocks were prepared. All experiments were performed after additional cell growth for several passages in the absence of selection in order to avoid physiological side effects of G418. To obtain optimal reproducibility, all transfectants were kept in culture for limited times only (fewer than 25 passages).
Western blotting analyses.
Protein extracts were separated by discontinuous SDS-10% PAGE and blotted onto nitrocellulose membranes. Proteins of interest were detected by incubation with appropriate primary antibodies in 10% dry milk-phosphate-buffered saline (PBS) for 18 h and staining with horseradish peroxidase-conjugated secondary antibodies for 1 h followed by chemiluminescence detection (Amersham).
Immunofluorescence microscopy.
For examination by immunofluorescence microscopy, cells were grown on spot slides, mock or MVMp infected, and further incubated for appropriate times. Cultures were fixed using 3% paraformaldehyde in PBS (pH 7.4) for 30 min at room temperature, neutralized with 50 mM NH4Cl in PBS for 6 min, and permeabilized by treatment with PBS containing 0.1% Triton X-100 for 10 min. All solutions were supplemented with 1 mM MgCl2 and 0.5 mM CaCl2. After extensive washing, cells were blocked with 10% goat serum for 30 min, incubated with primary antibodies for 2 h at room temperature, and last, incubated with fluorescein isothiocyanate (FITC)- or CY3-conjugated anti-species specific antibodies (Dianova) for detection purposes. After mounting using Elvanol, cells were analyzed by conventional epifluorescence microscopy (×63 lens with immersion oil; Leica) or confocal laser scanning microscopy (Leica TCS SP laser fitted to a Leica IMRBE microscope).
NS1 metabolic labeling, purification, and phosphopeptide analyses.
Metabolic labeling of NS1 and tryptic phosphopeptide analyses were essentially performed as previously described (43). A9 cell cultures (107 cells) were infected with MVMp (20 PFU/cell), incubated for 24 h before the labeling medium (complete medium lacking phosphate [Gibco/BRL] complemented with 0.1 nCi of [32P]orthophosphate [ICN]/cell) was added for 4 h. Labeled cells were harvested directly in 1 ml of RIPA buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Na-deoxycholate, 1% Triton X-100) containing protease and phosphatase inhibitors. NS1 immunoprecipitations were carried out using 10 μl of anti-NSN antiserum for 2 h at room temperature. Immune complexes were precipitated with protein A-Sepharose, washed with RIPA buffer, and further purified by SDS-10% PAGE. 32P-labeled proteins were revealed by autoradiography after blotting on polyvinylidene fluoride membranes, and the band corresponding to NS1 was excised. Digestion of membrane-bound NS1 was performed with 50 U of trypsin for 18 h at 37°C. Tryptic peptides contained in the supernatant were recovered by lyophilization and analyzed on thin-layer cellulose plates (Merck) in two dimensions, first by electrophoresis using a pH 1.9 buffer and then by chromatography in phospho-chromatography buffer.
MVM DNA replication in infected cells.
The accumulation of MVM DNA replicative forms was analyzed by Southern blotting as described by Corbau et al. (15). A9 cells (or derivatives A9:P38-PKCηA160E, A9:P38-PKCηT512A and A9:P38-PKCηDN) (3 × 105) were infected with MVMp (10 PFU/cell). Cells were harvested after 2, 24, 48, and 72 h postinfection in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and digested with proteinase K in 0.1% SDS for 18 h at 46°C. The whole-cellular DNA was then sheared by passage through a syringe, fractioned by 0.8% agarose gel electrophoreses, and blotted on a nitrocellulose membrane. Viral replicative intermediates were then detected using a 32P-labeled probe corresponding to nucleotides 385 to 1885 of the NS1-coding region of MVMp.
For in situ viral DNA replication studies, cells were grown on coverslips and infected with MVMp (50 PFU/cell) for 24 or 48 h. At various times, cells were labeled with 10 μM BrdU in DMEM for 25 min at 37°C and fixed with 3% paraformaldehyde. Replicating viral DNA was detected by immunofluorescence as described above using a monoclonal rat anti-BrdU antibody and quantitatively analyzed.
RESULTS
The novel isoform PKCη activates NS1 in cooperation with PKCλ for RCR.
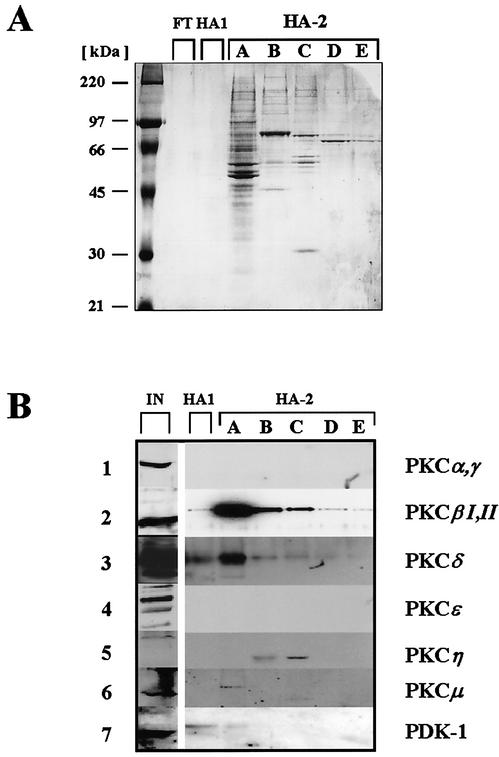
Previous investigations have identified atypical PKCλ phosphorylation to be essential to activate NS1 for initiation of viral DNA amplification. This phosphorylation event proved sufficient to activate the viral polypeptide for DNA unwinding during the initial nicking reaction and the processive helicase function in front of the replication fork (21, 42, 46). However, PKCλ phosphorylation alone did not enable dephosphorylated NS1O to drive RCR in concert with the cellular replication machinery (21, 45). Additional cellular factor(s) appear to be necessary, which due to their cofactor requirements (45) are also likely to belong to the PKC family. To identify such components involved in phosphorylation and activation of NS1 for RCR, we used a previously established complementation approach based on a kinase-free in vitro replication system (45). Circular plasmids containing the left-end origin of MVM DNA replication were subjected to RCR reactions in the presence of NS1O, recombinant PKCλ, and PKC-activating cofactors. In addition, the system was complemented with distinct fractions from HeLa cell extracts. Figure 1 depicts our purification strategy for identification of protein kinases, which activate NS1 for RCR in the presence of PKCλ. As previously described (45), the component(s) required to activate NS1O for RCR besides PKCλ copurified with PKCλ through phosphocellulose, strong anion exchange, and protamine affinity chromatography but could be separated from PKCλ due to their higher affinity to HA. Atypical PKCs, including PKCλ/ζ, were eluted from HA columns with as little as 20 mM KPO4 in the so-called HA-1 low-affinity fraction. The bound material constituting the high-affinity HA-2 fractions was previously shown to comprise a factor(s) complementing PKCλ (or HA-1) for NS1 activation in a TPA-dependent fashion (45). To characterize this factor(s), the HA-2 fraction was further purified by step-gradient elution. Five subfractions (A to E) eluting at 116, 164, 212, 260, and 500 mM KPO4, respectively, were collected. Interestingly, a significant accumulation of proteins in the range of 70 to 90 kDa was observed, particularly in fractions with higher HA affinity (Fig. 2A). This size, together with the previously reported cofactor requirements of HA-2 for phosphatidylserine and/or phorbol esters (such as TPA) in in vitro replication assays (45), prompted us to characterize individual subfractions eluting from HA columns by Western blotting for the presence of conventional and novel PKC isoforms. Figure 2B illustrates the distribution of individual PKC isoforms separating on HA. PKCα, PKCγ, and PKCɛ could not be detected in any HA fraction, since they became lost during previous chromatography steps. In contrast, PKCβI/II, PKCδ, PKCη, and PKCμ as well as the PKC activator kinase PDK-1 were retained on HA due to their distinct affinity to the matrixes used in the purification procedure. The consecutive elution of the HA-bound protein kinases with increasing amounts of KPO4 revealed the PKC isoforms PKCβI/II, PKCδ, PKCμ, and PDK-1 to be recovered at relatively low concentrations, with the greater part of each of these proteins being present in subfraction HA-2A. Interestingly, although peak amounts of these kinases were indeed released in HA-2, a significant proportion was still present in HA-1 (Fig. 2B, lane 2), hence excluding PKCβI/II, PKCδ, PKCμ, and PDK-1 as coactivators for atypical PKCλ (21, 45). In contrast, the novel PKCη was retained on the HA matrix during the first elution steps, coming off the column at a rather high KPO4 concentration in subfraction C, with only a minor proportion being present in subfraction HA-2B.
FIG. 2.
Analysis of the proteins present in HA-2 subfractions. (A) Whole protein staining. A sample of each fraction was analyzed by SDS-10% PAGE, followed by fixing and staining with Coomassie brilliant blue. FT, flowthrough (HA-0); HA1, HA fraction 1 eluted with 20 mM KPO4 and containing PKCλ/ζ; HA-2, HA fraction 2 eluted at KPO4 concentrations ranging from 116 to 500 mM. All fractions were enriched in proteins of 70 to 90 kDa, corresponding to the sizes of PKC isoforms. (B) Western blot detection of distinct protein kinases. HeLa cell extracts and HA subfractions separated by SDS-PAGE were analyzed for the presence of indicated protein kinases by Western blotting using isoform-specific antibodies.
The HA-2 subfractions were then tested for their capacity to activate dephosphorylated NS1O for RCR initiation in a kinase-free in vitro replication system. This in vitro assay is based on a Thr-affinity-purified subcellular fraction containing RPA, PCNA, the parvovirus initiation factor PIF, and bacteriophage T4 DNA polymerase. To search for the additional NS1-activating factors present in HA-2 (subfractions), we supplemented the replication reactions with the PKC cofactors PS and TPA as well as recombinant PKCλ to activate NS1 DNA unwinding functions (21, 42). All replication reactions were performed at 37°C for 2 h in the presence of [α-32P]dATP. DNA was then immunoprecipitated with anti-NSN and analyzed by 0.8% agarose gel electrophoresis in order to discriminate between random nick-translation activity and genuine NS1-initiated replication products. As illustrated in Fig. 3 (lanes 3 to 9), no detectable RCR activity was obtained with dephosphorylated NS1O in the absence of PKC and, as expected from previously published results (21, 46), only a marginal stimulation was observed upon addition of recombinant PKCλ alone (lane 5). In contrast, further supplementation of the assay with distinct HA-2 subfractions eluting at high KPO4 concentrations resulted in a significant rescue of NS1O replication activity. Maximum rescue was achieved with fraction HA-2C, which cooperated with PKCλ to activate NS1O up to a 10-fold-higher level than PKCλ alone (Fig. 3, lane 7). Reactivation of NS1O dropped significantly towards both ends, elution with higher and lower KPO4 concentrations, being only 5- and 2-fold for HA-2D and HA-2B, respectively (Fig. 3, lanes 6 and 8) and getting undetectable for HA-2A (data not shown). This distribution allowed us to draw a parallel between the NS1 activation potential of the different HA-2 subfractions and their respective contents in PKC as measured by Western blotting. Among the kinases detected within the HA-2 components, only PKCη was found to concentrate in the most active HA-2C subfraction (compare Fig. 2B and 3). In contrast, the elution of PKCβI/II, PKCμ, or PKCδ peaking in HA-2A did not match the activation profile, while PKCα, PKCγ, and PKCɛ were already excluded as candidate coactivators from prior purification steps. Thus, this correlation strongly argued for an involvement of the novel PKCη isoform in the activation of NS1 for RCR activity.
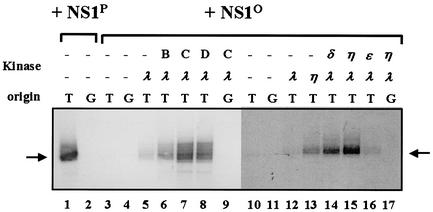
FIG. 3.
Reactivation of dephosphorylated NS1 in in vitro replication assays. NS1-dependent RCR of plasmids containing the left-end active (T) or inactive (G) origin was determined in a kinase-free in vitro system based on P1-Thr and T4 DNA polymerase (45). The reaction products were analyzed by 0.8% agarose gel electrophoresis after immunoprecipitation with anti-NSN antiserum, HindIII restriction digestion, and deproteination. The migration of the linearized plasmid is indicated with an arrow. Lanes 1 and 2, fully phosphorylated NS1; lanes 4 to 17, dephosphorylated NS1O using alkaline phosphatase; lanes 5 to 9 and 12 to 17, reactions performed in the presence of indicated HA-2 subfractions (B, C, and D) and/or recombinant activated PKCs (λ, η, δ, and ɛ).
In order to determine whether PKCη is indeed able to activate NS1 for RCR, we carried out the in vitro replication reaction in the presence of recombinant PKCλ and PKCη without the additional components present within the HA-2 subfractions. To produce recombinant PKCη, we isolated a full-length cDNA clone from a human placenta cDNA library, cloned it into plasmid pTHis1, and generated recombinant vaccinia viruses. Expression of PKCη was driven under control of bacteriophage T7 polymerase and an encephalomyocarditis virus leader sequence for cap-independent high efficiency translation in the presence of a second recombinant vaccinia virus, vTF7-3, to provide the T7 RNA polymerase. The recombinant protein was produced in HeLa cells and purified by means of its N-terminal His6 tag on Ni2+-nitrilotriacetic acid agarose columns. For comparison, we also generated recombinant vaccinia viruses expressing the novel PKCδ and PKCɛ, since these closely related kinases might be substituted for PKCη under in vitro conditions when applied at high concentrations. The impact of these PKCs on NS1 regulation was then tested in in vitro replication assays as described above, replacing the HA-2 subfractions with the different recombinant PKC isoforms. As shown in Fig. 3 (lanes 10 to 17), recombinant PKCη was indeed able to stimulate the NS1O replication activity more than 10-fold in the presence of PKCλ, while PKCη alone was inefficient (lane 13). PKCɛ (lane 16) had only a marginal effect, stimulating NS1O by two- to threefold in comparison with PKCλ alone. Interestingly, recombinant PKCδ expressed in mammalian cells was able to substitute for PKCη in NS1O rescue, but with a significantly lesser efficiency (∼fivefold increase of activation achieved by PKCλ alone), in keeping with the somewhat relaxed phosphorylation specificity of PKCs under in vitro conditions (38). No activation was obtained by complementing PKCλ with recombinant conventional PKCs (PKCα, PKCγ, and PKCβI/II) or PKCδ expressed in insect cells from recombinant baculoviruses (data not shown). In summary, although high amounts of purified recombinant PKCδ were able to stimulate to some extent the in vitro replication activity of NS1O in the presence of PKCλ, our results obtained with fractionated cell extracts and recombinant PKCs provide strong evidence to suggest that PKCη is a prime candidate for the regulation of NS1 replicative functions in cooperation with PKCλ.
Production of A9 cell-derived stable transfectants harboring MVM-inducible PKCη mutant clones.
In order to assess a possible role of PKCη during MVM infection, we chose to modulate PKCη activity in A9 cells through the expression of mutant forms of the enzyme. For this purpose, we stably transfected A9 cell lines with PKCη mutant clones under control of the NS1-inducible P38 promoter. The generation of such stable transfectants offers several advantages. (i) As determined with a P38-driven EGFP construct (pP38-GFP), more than 95% of stably transfected cells were able to produce the foreign polypeptide upon infection, with no need to use other inducers that may perturb the system (data not shown). (ii) Due to integration into the chromosomal background, expression levels remained in a physiological range even upon induction for the majority of cells. Quite often a controlled expression of a foreign sequence can be problematic by episomal expression in transient-transfection assays (see below). (iii) Since the P38 promoter is induced through the viral protein NS1, elevated expression of the transgene should be reached after establishment of the MVM infection and, therefore, should have little impact on receptor interaction or virus entry. (iv) The introduction into the chromosomal background and steady-state expression of the transgene do not affect viability and propagation of the cell population, since they have to be maintained over an extended time period during the selection procedure.
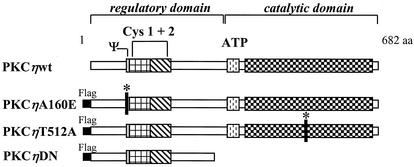
As illustrated in Fig. 4, the following PKCη mutants were constructed to alter endogenous PKCη activity. By mutagenesis of the pseudosubstrate domain at amino acid position 160 from alanine to glutamic acid, we generated the constitutively active variant PKCηA160E. It has been demonstrated for other PKC isoenzymes (4; J. P. F. Nüesch, unpublished data) that similar mutations at this site render PKC isoforms' activity independent of cofactors such as phosphatidylserine, diacylglycerol, or TPA due to reduced intramolecular binding of the regulatory domain to the substrate recognition site in the catalytic pocket. In contrast, substitution of alanine for threonine at T512, a PDK-phosphorylation site within the activation loop, results in a catalytically inactive mutant (PKCηT512A). Indeed, this PKCη variant does not undergo the phosphorylation-dependent conformational changes required for its functioning, since the T512A substitution prevents the kinase from all (auto)phosphorylations at its C terminus (23; Nüesch, unpublished data). Overexpression of the mutant form PKCηT512A is expected to inhibit endogenous PKCη activity due to substrate competition. In addition, we constructed a dominant-negative mutant, PKCηDN, consisting of the isolated regulatory domain. As previously established for PKCλ (46), overexpression of the regulatory domain (aa 1 to 296) inhibits endogenous active PKC. It is thought that this polypeptide, PKCηDN, which harbors the pseudosubstrate site, blocks the substrate recognition motif of PKCη with high specificity and affinity. Since this artificially expressed regulatory domain is not connected to the kinase, conformational alterations induced by cofactor interactions are not able to release this block of the active site (39). All three mutants were constructed with an N-terminal Flag epitope, cloned into the expression plasmid pP38 and stably transfected into the mouse fibroblast cell line A9, in the presence of a neomycin resistance plasmid, pSV2neo.
FIG. 4.
Construction of PKCη mutants. PKCη is a member of the novel PKC family consisting of 682 aa. Besides the catalytic domain comprising the substrate recognition and ATP-binding sites, PKCs contain a large regulatory domain. This domain is characterized by the presence of a pseudosubstrate sequence (Ψ) and two cysteine-rich Zn finger motifs (Cys 1 + 2) binding the cofactors phosphatidylserine, DAG, and TPA. PKCηA160E is a constitutively active mutant form of human PKCη in which the pseudosubstrate region was altered to prevent it from binding to the catalytic domain, keeping the enzyme in an open conformation. The inactive mutant PKCηT512A was obtained by an amino acid substitution for the PDK1 phosphorylation site T512, mimicking the nonphosphorylated residue. The dominant negative mutant PKCηDN was constructed by deleting the C terminus from aa 297 onwards and should block the substrate recognition site of endogenous PKCη through high-affinity binding. A Flag epitope was N-terminally fused to all PKCη mutants, enabling their distinction from the endogenous enzyme through immunodetection.
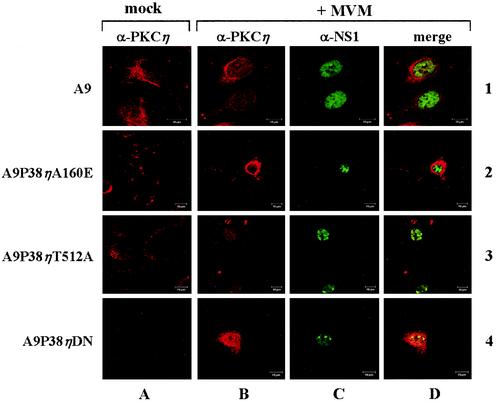
The isolated stable transfectants were first analyzed by confocal immunofluorescence microscopy to determine expression and subcellular localization of the recombinant proteins (PKCηA160E, PKCηDN, and PKCηT512A) in comparison to the endogenous PKCη in the parental A9 cell line. Cells were seeded on spot-slides, infected or not with 20 PFU of MVMp/cell, and fixed with 3% paraformaldehyde 24 h postinfection. Typical immunofluorescence data of PKCη and NS1 are shown in Fig. 5. Endogenous PKCη was located throughout the cytoplasm of noninfected A9 cells in association with filamentous structures (panel 1A). Upon MVM infection, PKCη remained cytoplasmic but showed a clear perinuclear accumulation in the majority (50 to 60%) of NS1-expressing cells (panels 1B to 1D). According to the current model of PKC activation cascades, this redistribution could indicate activation of the kinase as a result of MVM infection (39). Recombinant PKCη variants could be detected independently of the endogenous protein due to their N-terminal Flag epitope (Fig. 5, lanes 2 to 4). As expected from previous analyses using episomal (53) or stably integrated P38 promoter constructs (41), expression of the PKCη variants was stimulated upon MVM infection in our stable cell lines, though to an extent which varied within the cell population (rows B and D versus row A). These variations are most likely due to the fact that pooled, rather than cloned, transfectants were used in our experiments, resulting in heterogeneous transgene integration patterns.
FIG. 5.
Confocal immunofluorescence detection of PKCη expressed from the endogenous gene and transfected mutant clones. A9 cells (lane 1) or stably transfected derivatives thereof (lanes 2 to 4) were grown on spot slides, infected (rows B to D) or not (row A) with MVMp (20 PFU/cell), and fixed at 44 h p.i. Endogenous PKCη was detected with a polyclonal rabbit antiserum and CY3-labeled anti-rabbit secondary antibodies, while the mutant PKCη proteins were visualized with a mouse monoclonal Flag antibody and CY3-conjugated anti-mouse secondary antibody. NS1 was revealed using either a monoclonal mouse antibody and FITC-conjugated secondary anti-mouse antibodies or polyclonal rabbit antiserum and FITC-labeled anti-rabbit antibodies. All images were acquired using a Leica TCS SP Microscope (×63 oil immersion lens, FITC/tetramethyl rhodamine isocyanate filter settings, and a pinhole of 1). Cells were stained for endogenous PKCη (panels 1 A and B), recombinant PKCη (panels 2A and B to 4A and B), or NS1 (panels 1 to 4C); panel D, merged images.
The recombinant full-length PKC variants showed a uniform cytoplasmic distribution similar to that of the endogenous polypeptide. Some unique features of this distribution are worth mentioning. Although the majority of the population expressed the transgene at levels close to the detection limit, approximately 10% of the cells showed strong accumulation of the PKCη variants upon MVM infection. Within these overexpressing cells, the constitutively active PKCηA160E and the catalytically inactive PKCηT512A were both found to have a cytoplasmic localization (Fig. 4, lanes 2 and 3), suggesting that PKCη regulation may not involve the nuclear translocation as previously reported for classical PKCs (39). The expression level of the N-terminal part of PKCη (the dominant-negative PKCηDN) was very low even upon induction by MVM NS1 (Fig. 5, panels 4B to 4D), with the exception of a few strongly overexpressing cells, as shown in Fig. 5 (panel 4B). In contrast to the full-length enzymes, this polypeptide was found in the cytoplasm, nucleus, or vicinity of the nuclear membrane, probably as a result of its relatively small size (<40 kDa). Altogether, these data showed that the recombinant PKCη variants were indeed expressed upon infection with MVM and constituted a suitable tool to be tested for their effects on virus replication.
The production of all recombinant proteins was also confirmed through Western blotting and in vitro transcription-translation experiments. A rather high basal level of expression of PKCηA160E and PKCηT512A under control of the P38 promoter was detected even under noninduced conditions. However, this expression level was significantly increased (between twofold [for PKCηT512A] and fivefold [for PKCηA160E]) upon MVM infection (data not shown). In addition, it should be stated here that the cell populations harboring either one of the three PKCη constructs were competent for MVMp entry and expression of NS1 proteins (Fig. 5, row C). Yet the efficiency of virus uptake, as measured by the presence of single-stranded DNA (Southern blot) or capsids (immunofluorescence) at different time points postinfection (p.i.), was significantly impaired under PKCη knock-down conditions. Indeed, it appeared that the virus uptake was delayed for several hours in the presence of PKCηDN or ηΤ512A (data not shown). Despite these side effects, the cell lines expressing PKCη variants are still valuable tools for studying the impact of PKCη on MVM DNA replication in vivo.
NS1 phosphorylation by PKCη in vitro and in vivo.
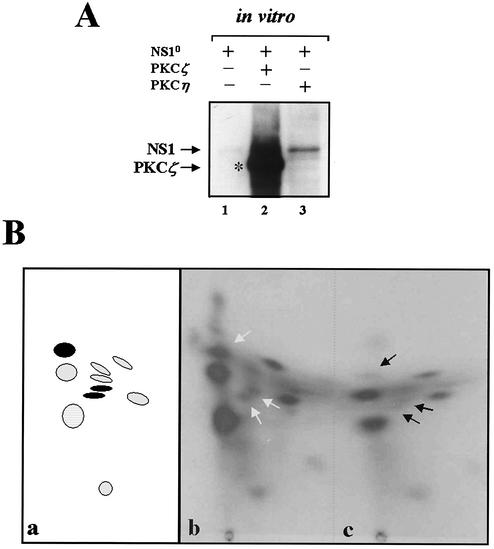
The above-mentioned activation of dephosphorylated NS1O for replication activity in vitro by recombinant PKCη provides strong evidence to suggest that NS1 constitutes a direct substrate for phosphorylation and regulation by PKCη. To further study the NS1 PKCη interplay, we investigated whether PKCη is indeed able to phosphorylate NS1 in vitro, and if so, whether NS1 is a substrate for this protein kinase in vivo as well. In vitro phosphorylation of dephosphorylated NS1O was first tested in the presence of the PKC activators PS and TPA, [γ-32P]ATP, and recombinant PKCη. As a positive control the assay was also carried out using recombinant PKCζ, also derived from vaccinia virus expression in HeLa cells, for which NS1O is known to be a target in vitro (21). The 32P-labeled NS1 protein was then immunoprecipitated with anti-NS1H and analyzed by SDS-PAGE. Although the extent of phosphorylation achieved by PKCη was significantly less than that with PKCζ, NS1O proved to be a target substrate for PKCη in vitro (Fig. 6A, lanes 1 to 3). This result shows that NS1 indeed serves as a direct substrate for PKCη phosphorylation in vitro. Therefore, this kinase might also act as a direct regulator of NS1-driven replication in vivo, as suggested by the PKCη-mediated activation of NS1O for RCR in in vitro assays.
FIG. 6.
Phosphorylation of NS1 by PKCη under in vitro and in vivo conditions. For in vitro phosphorylation experiments (A), dephosphorylated NS1O was incubated with purified recombinant PKCη or PKCζ in the presence of [γ-32P]ATP and the PKC activators TPA and PS. For in vivo phosphorylation experiments, A9 or A9:P38-PKCηDN cells were infected with 30 PFU of MVMp/cell and labeled with [32P]orthophosphate for 4 h at 24 h p.i. 32P-labeled NS1 proteins were isolated by immunoprecipitation and processed for further analyses. (A) One-dimensional SDS-PAGE of NS1 proteins that were phosphorylated in vitro by incubation with the kinases indicated on top (lanes 1 to 3). The migration of NS1 and that of the coprecipitated, autophosphorylated PKCζ are indicated. (B) Tryptic phosphopeptide analysis of metabolically 32P-labeled NS1 proteins derived from MVM-infected A9 (b) or A9: P38-PKCηDN (c) cells. Arrows point at phosphopeptides that are suppressed in the presence of the PKCη dominant-negative form. In the left-hand scheme (a), the peptides that are targets for PKCη (present study) and PKCλ (46) are marked in black and hatched, respectively.
To determine whether PKCη also contributes to NS1 phosphorylation in the cellular environment, we performed metabolic 32P labeling and tryptic phosphopeptide mapping of purified NS1 proteins after MVM infection. To inhibit the endogenous PKCη activity, we used the P38-PKCηDN cell line and compared the phosphorylation pattern of NS1 in these cells and the parental A9s during the replicative phase of an MVM infection. Subconfluent A9 or A9:P38-PKCηDN cells were infected with 30 PFU of MVMp/cell for 20 h and metabolically 32P labeled for an additional 4 h. Cells were harvested directly into lysis buffer 24 h postinfection, and NS1 was isolated by immunoprecipitation using anti-NS1N antiserum and further purified by SDS-PAGE. The NS1-associated phospho-labeling was much reduced in the presence of PKCηDN (data not shown). This overall effect could be caused by impairment of NS1 phosphorylation and/or production. In order to distinguish these two possibilities and to determine whether specific NS1 residues were indeed targeted by PKCη, we performed two-dimensional tryptic phosphopeptide analyses. The loss or reduction of a distinct NS1 phosphopeptide(s) under PKCη knock-down conditions should indeed provide direct evidence of the involvement of PKCη in NS1 phosphorylation. As previously reported (15, 43) and illustrated in Fig. 6B (a and b), NS1 from MVM-infected A9 cells (panel b) showed a characteristic tryptic phosphopeptide pattern after two-dimensional thin-layer electrophoresis-chromatography. This overall pattern of NS1, including the characteristic PKCλ phosphopeptides, could still be recognized under conditions where endogenous PKCη was inhibited by the presence of PKCηDN. However, in contrast to the A9-derived phosphopeptide pattern, three distinct phosphopeptides (Fig. 6B, panel c, arrows) were markedly underrepresented. This selective inhibition of distinct phosphorylation events in the presence of PKCηDN argues for the contribution of PKCη in the phosphorylation of NS1 in A9 cells. Moreover, the involvement of PKCη in the phosphorylation of NS1 in vivo is in agreement with the proposed role of this cellular kinase for the regulation of NS1 functioning.
Impact of PKCη phosphorylation of NS1 on MVM DNA replication.
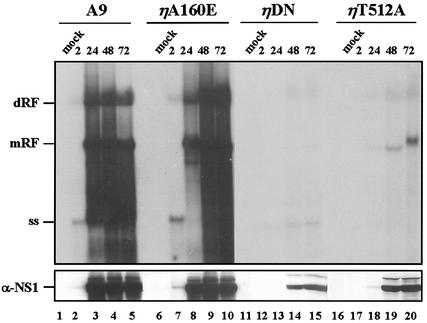
The results described above using the kinase-free in vitro replication system indicate that PKCη phosphorylation is essential to enable NS1O (in concert with PKCλ) to drive viral DNA amplification at least in vitro. To ascertain whether phosphorylation of NS1 by PKCη was indeed of functional relevance in vivo, we set out to analyze MVM DNA replication in the presence of the PKCη variants, i.e., under conditions modulating the intracellular PKCη activity. For this purpose the previously described stably transfected A9 derivatives (A9:P38-PKCηA160E, A9:P38-PKCηT512A, and A9:P38-PKCηDN) were used and analyzed for their competence in viral DNA amplification in comparison with the parental A9 cells in time-course experiments. As illustrated in Fig. 7 (upper panel, lanes 1 to 10), the parental A9 cell line and A9:P38-PKCηA160E (overexpressing the constitutively active PKCη mutant form) were both able to support the production of viral DNA replicative intermediates (monomeric and dimeric RF) as well as progeny single-strand virion DNA to similar amounts at all time points. In contrast, A9:P38-PKCηT512A and A9:P38-PKCηDN cells were both impaired to support viral DNA amplification. In fact, even as late as 72 h postinfection (lanes 11 to 20), viral replication intermediates were barely detectable under these conditions. Thus, inhibiting endogenous PKCη activity resulted in a marked suppression of the cell capacity for MVM DNA replication. Although delayed virus entry was observed for these PKCη cell lines, it cannot account for the dramatic effect on viral DNA amplification observed with A9:P38-PKCηDN and A9:P38-PKCηT512A, particularly since the amount of replicative forms does not increase even as late as 72 h p.i. In addition, the poor viral DNA amplification in the presence of PKCηDN or PKCηT512A also caused a striking reduction in the production of viral proteins, as shown for NS1, presumably due to limiting amounts of transcription templates (Fig. 7, lower panel). Such loop-back regulation could certainly contribute significantly to the dramatic negative effect observed on the replication activity under these conditions.
FIG. 7.
MVM DNA replication and NS1 protein production in A9 cells and transfectants overexpressing mutant forms of PKCη. A9 cells, or derivatives stably transfected with indicated P38-driven PKCη mutant clones, were infected or not with MVMp (10 PFU/cell) and harvested at various time points (hours p.i.). Viral DNA and NS1 proteins were quantitated by Southern (upper panel) and Western blotting (lower panel), respectively. mRF, monomeric replicative intermediate form; dRF, dimeric replicative intermediate form; ss, single-stranded DNA.
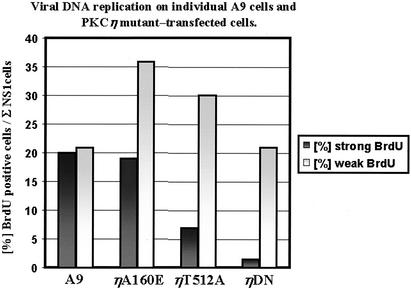
To rule out that catalytic inactive PKCη mutants decreased the overall viral DNA amplification merely due to a reduced fraction of cells that became infected, MVM DNA replication was also measured on the single-cell level. For this purpose, A9, A9:P38-PKCηA160E, A9:P38-PKCηT512A, or A9:P38-PKCηDN cells were seeded on coverslips and infected with MVMp (multiplicity of infection, 50). Either 24 or 48 h postinfection, the culture medium was replaced with 10% DMEM containing 100 nM BrdU to allow incorporation of the nucleotide for 25 min into replicating DNA. To determine the rate of viral DNA replication, cells were fixed with 3% p-formaldehyde and subjected to double-immunofluorescence analysis using monoclonal rat anti-BrdU and polyclonal rabbit anti-NS1 antiserum. In the absence of HCl treatment, incorporation of BrdU into replicating chromosomal DNA cannot be detected (2, 61). This allowed the generation of parvoviral replicative forms during the labeling period to be revealed by anti-BrdU immunostaining. Only NS1-positive cells were analyzed for the extent of BrdU incorporation, to rule out side effects of the PCKη knock-down prior to the establishment of a productive infection. Since the intensity of BrdU staining varied significantly between individual cells in an infected population, we decided to distinguish between weak (punctate) and strong (full-blown) BrdU signals. The results obtained 24 and 48 h p.i. are summarized in Fig. 8. As expected from our analyses of the whole-cell population by Southern blotting, the cell lines with reduced PKCη activity could be distinguished from the parental and constitutively active A9:P38-PKCηA160E cell line by a striking reduction of the fraction of NS1-positive cells that also scored positive for strong BrdU incorporation. Inactivation of PKCη in the A9:P38-PKCηT512A and A9:P38-PKCηDN cell lines did not allow virus replication to recover at later time points. On the contrary, the fraction of NS1-positive cells with strong BrdU signals dropped further with time. Since only NS1-positive cells, i.e., cells that were able to establish a productive infection, were taken into account, these results confirm that conditions inhibiting the endogenous PKCη activity also impaired the capacity of cells to support viral DNA amplification, irrespective of their competence of viral uptake. Together with the Southern blotting and in vivo phosphorylation analyses, these data strongly support the functional role of PKCη in the regulation of NS1 during viral DNA amplification.
FIG. 8.
Quantitative evaluation of MVM DNA replication activity in A9 cells and derivatives overexpressing mutant forms of PKCη at the single-cell level. Cells infected with MVMp (50 PFU/cell) were pulsed with BrdU at 1 or 2 days p.i. and tested for incorporation of BrdU into replicating viral DNA. Only cells showing a positive NS1 signal were analyzed. NS1 was detected using specific polyclonal rabbit antiserum and FITC-conjugated anti-rabbit secondary antibodies. BrdU labeling was monitored by immunostaining with rat monoclonal anti-BrdU antibody and Alexa Fluor 555-conjugated anti-rat secondary antibodies. Punctuated and full-blown nuclear BrdU signals were defined as weak and strong, respectively. Columns represent the sum percentage of BrdU per NS1-positive cells from both days.
DISCUSSION
Previous investigations have shown that MVM DNA amplification is tightly controlled by phosphorylation of the initiator protein NS1 (43, 45). Extensive in vitro analyses using site-directed mutants with defined biochemical profiles have identified PKCλ as an essential protein kinase which controls the initiation of viral DNA replication and strand-displacement synthesis through regulation of the NS1 DNA unwinding activities (21, 42). However, while PKCλ phosphorylation of NS1O is sufficient to enable the initial nicking reaction, subsequent DNA synthesis controlled by NS1 requires additional phosphorylation events (42, 46). The data presented here identify an additional member of the PKC family, the novel isoform PKCη, which is able to phosphorylate and regulate the large nonstructural protein NS1 of MVMp. We show that PKCη activates NS1 in cooperation with PKCλ for RCR in vitro. This in vitro activation appears to be specific for PKCη, since other members of the PKC family, such as the conventional PKCα, PKCβI/βII, and PKCγ or the novel PKCɛ, failed to activate NS1, despite their ability to phosphorylate the viral polypeptide (21). To study the impact of PKCη on viral DNA replication in vivo, we produced stably transfected cell lines whose overall PKCη activity was altered due to the expression of mutant forms of this kinase. Inhibition of endogenous PKCη activity upon MVM infection led to a drastic reduction in the accumulation of viral DNA replication intermediates as measured in the whole-cell population as well as on the level of the single cell. Together with the observed alterations of the NS1 phosphorylation pattern in the presence of a dominant-negative PKCη mutant, these results underline the importance of PKCη for the regulation of a productive MVM infection in permissive cells.
The mechanism by which PKCη controls NS1-driven MVM DNA amplification remains to be elucidated. Unwinding and nicking of the left-end origin under physiological conditions could be reconstituted in vitro using dephosphorylated NS1 and activated purified recombinant PKCλ (42). This finding demonstrates that PKCη is dispensable for the activity of a number of NS1 functions, such as ATP binding and hydrolysis, oligomerization, site-specific interaction with the cognate DNA motif, cleavage of the single-stranded nicking motif, trans esterification with the free 5′ end of nicked DNA, and the interaction with the cellular cofactor PIF. Furthermore, the processive helicase function of NS1, which is thought to act in front of the replication fork to promote strand-displacement synthesis achieved by polymerase δ, is independent of PKCη phosphorylation (21). This leads us to speculate that PKCη may regulate the interaction of NS1 with the cellular DNA replication machinery, coupling the opening of duplex viral DNA intermediates with leading strand synthesis. NS1 has recently been shown to interact with various components of the replication complex, including the single-strand binding protein RPA and the accessory protein complex RF-C (10). Altogether, these and the present observations raise the intriguing possibility that the interaction of NS1 with one and/or another of these factors (including the active polymerase) might be regulated by PKCη phosphorylation of the viral products. Indeed, phosphorylation can control protein-protein interactions, since the transfer of a negative charge to the protein is often accompanied with a change in the conformation of the polypeptide and, in consequence, its affinity towards distinct partner proteins (38). Identification of the target PKCη phosphorylation sites in NS1, assignment of these sites to functional domains, and molecular analyses of the corresponding NS1 mutants should help to further characterize the NS1 feature(s), which is (are) regulated through PKCη.
Although inhibition of endogenous PKCη mainly affected viral DNA amplification, virus uptake was also impaired to a significant extent in the presence of kinase-inactive PKCη mutants. In contrast, the cell line expressing the constitutively active mutant PKCηA160E was fully proficient in this process. The mechanism by which PKCη may regulate virus entry is currently a matter of speculation, especially since the MVM receptor remains to be identified. It is noteworthy, however, that there are precedents for an impact of PKCs on receptor-activated processes. Indeed, PKC activity was found to correlate with the amount of the retinoic acid receptor protein (8), to affect internalization of some receptors (6) including the serotonin receptor 5-HT2A (7), to regulate the intracellular trafficking of the insulin growth factor receptor II (29), or to promote receptor recycling (5, 57). In contrast, the transferrin receptor, which has been identified as the receptor for the canine parvovirus (51), was shown to be downregulated upon prolonged PKC activation by phorbol esters (55). Altogether, considering these observations, it seems conceivable that under reduced PKCη activity (as achieved in the presence of the inactive PKC mutants) the MVM receptor might become a limiting factor, due either to its amount or to its ability to guide the intracellular trafficking of the input virus.
The propagation of autonomous parvoviruses is highly dependent upon the proliferation and differentiation state of host cells. These restrictions are likely to account for the preferential multiplication of these agents in a number of neoplastically transformed cells, a property designated oncotropism (for a review, see reference 54). Given their disregulation in many transformed cells, protein kinases, particularly PKCs, are intriguing candidates for oncotropic determinants to stimulate parvovirus propagation in these cells. It is worth noting in this respect that in contrast to the ubiquitous PKCλ, PKCη is expressed in a tissue-specific fashion. The modulation of PKCη by differentiation can be exemplified by predominant expression in cells of epithelial origin (3, 26, 50) and in neuronal precursors (24, 47). Furthermore, like other PKC isoforms, PKCη is activated in the presence of tumor promoters, such as TPA. It is therefore possible that neoplastic transformation of certain tissues may result in activation of PKCη, which contributes to their enhanced permissiveness for parvovirus infection. This putative role of PKCη in cell tropism of parvoviruses remains, however, to be tested experimentally.
PKC family members often undergo intracellular translocation upon activation, becoming increasingly bound to insoluble membranes as demonstrated by immunofluorescence and biochemical analyses (39). In unstimulated endothelial cells, for instance, PKCη was found predominantly in the perinuclear region or at the Golgi apparatus, while induction through TPA led to the translocation of a significant proportion of these enzymes towards the nuclear membrane and into the nucleus (27). Our immunofluorescence analyses of A9 fibroblast cells showed a similar redistribution of endogenous PKCη upon MVM infection, although PKCη was unable to enter the nucleus even as a constitutively active variant. In summary, despite the fact that our observations need additional investigations, the translocation of endogenous PKCη upon MVM infection provides the first evidence to suggest that viral infection is indeed accompanied by activation of this kinase in A9 cells.
As a result of their subcellular redistribution and activation (48) and their interaction with distinct cofactors or partner proteins (30), PKC isoforms are able to induce a wide spectrum of signaling cascades (56, 59). This makes it possible that a single PKC isoform is able to take part in multiple (and even antagonistic) signaling events, depending on the cell type and/or the trigger event involved. The redistribution of PKCη induced by MVM is likely to give rise to a positive feedback loop, causing the activated enzyme to stimulate MVM DNA replication through NS1 modification. In addition, however, the activated kinase may also conceivably serve different functions advantageous for the virus life cycle. Stimulation of PKC isoenzymes, (including the novel PKCη and PKCδ) have indeed been shown to have a strong impact on cell physiology, influencing more particularly cell cycle progression (25, 37), differentiation, and transformation (31, 32). In agreement with the observed morphological alterations imposed on the host cell upon infection and overexpression of PKCη, this interplay with the PKC signaling pathway could ensure a suitable environment for virus propagation after infection of target cells.
Acknowledgments
Particular thanks are given to Claudia Plotzky for excellent technical assistance. We are also indebted to Bernard Moss (NIH) for making the T7-driven vaccinia virus expression system available to us. In addition, we are grateful to Peter Tattersall and Susan Cotmore for providing us with multiple plasmid constructs and antisera.
REFERENCES
- 1.Anouja, F., R. Wattiez, S. Mousset, and P. Caillet-Fauquet. 1997. The cytotoxicity of the parvovirus minute virus of mice nonstructural protein NS1 is related to changes in the synthesis and phosphorylation of cell proteins. J. Virol. 71:4671-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, A., S. Shibui, M. Barker, M. Vanderlaan, J. W. Gray, and T. Hoshino. 1990. Cell kinetics of rat 9L brain tumors determined by double labeling with iodo- and bromodeoxyuridine. J. Neurosurg. 73:254-258. [DOI] [PubMed] [Google Scholar]
- 3.Bacher, N., Y. Zisman, E. Berent, and E. Livneh. 1991. Isolation and characterization of PKC-L, a new member of the protein kinase C-related gene family specifically expressed in lung, skin, and heart. Mol. Cell. Biol. 11:126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baier-Bitterlich, G., F. Uberall, B. Bauer, F. Fresser, H. Wachter, H. Grunicke, G. Utermann, A. Altman, and G. Baier. 1996. Protein kinase C-theta isozyme selective stimulation of the transcription factor complex AP-1 in T lymphocytes. Mol. Cell. Biol. 16:1842-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao, J., I. Alroy, H. Waterman, E. D. Schejter, C. Brodie, J. Gruenberg, and Y. Yarden. 2000. Threonine phosphorylation diverts internalized epidermal growth factor receptors from a degradative pathway to the recycling endosome. J. Biol. Chem. 275:26178-26186. [DOI] [PubMed] [Google Scholar]
- 6.Beeler, J. F., and R. H. Cooper. 1995. Regulation of hepatocyte plasma membrane alpha 1-adrenergic receptors by 4 beta-phorbol 12-myristate 13-acetate. Biochem. J. 305:73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharyya, S., S. Puri, R. Miledi, and M. M. Panicker. 2002. Internalization and recycling of 5-HT2A receptors activated by serotonin and protein kinase C-mediated mechanisms. Proc. Natl. Acad. Sci. USA 99:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boskovic, G., D. Desai, and R. M. Niles. 2002. Regulation of retinoic acid receptor alpha by protein kinase C in B16 mouse melanoma cells. J. Biol. Chem. 277:26113-26119. [DOI] [PubMed] [Google Scholar]
- 9.Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 9:2989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, J., S. F. Cotmore, and P. Tattersall. 1997. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J. Virol. 71:1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen, J., M. Pedersen, B. Aasted, and S. Alexandersen. 1995. Purification and characterization of the major nonstructural protein (NS-1) of Aleutian mink disease parvovirus. J. Virol. 69:1802-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen, J., S. F. Cotmore, and P. Tattersall. 1995. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 69:5422-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbau, R., V. Duverger, J. Rommelaere, and J. P. F. Nüesch. 2000. Regulation of MVM NS1 by protein kinase C: impact of mutagenesis at consensus phosphorylation sites on replicative functions and cytopathic effects. Virology 278:151-167. [DOI] [PubMed] [Google Scholar]
- 15.Corbau, R., N. Salomé, J. Rommelaere, and J. P. F. Nüesch. 1999. Phosphorylation of the viral non-structural protein NS1 during MVMp infection of A9 cells. Virology 259:402-415. [DOI] [PubMed] [Google Scholar]
- 16.Cotmore, S. F., and P. Tattersall. 1994. An asymmetric nucleotide in the parvoviral 3′hairpin directs segregation of a single active origin of DNA replication. EMBO J. 13:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotmore, S. F., and P. Tattersall. 1995. DNA replication in the autonomous parvoviruses. Semin. Virol. 6:271-281. [Google Scholar]
- 18.Cotmore, S. F., and P. Tattersall. 1998. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J. Virol. 72:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotmore, S. F., J. Christensen, J. P. F. Nüesch, and P. Tattersall. 1995. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2-3. J. Virol. 69:1652-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cziepluch, C., E. Kordes, R. Poirey, A. Grewenig, J. Rommelaere, and J.-C. Jauniaux. 1998. Identification of a novel cellular TPR-containing protein, SGT, that interracts with the nonstructural protein NS1 of parvovirus H-1. J. Virol. 72:4149-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dettwiler, S., J. Rommelaere, and J. P. F. Nüesch. 1999. DNA unwinding functions of minute virus of mice NS1 protein are modulated by the lambda isoform of protein kinase C. J. Virol. 73:7410-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doerig, C., B. Hirt, J. P. Antonietti, and P. Beard. 1990. Nonstructural protein of parvovirus B19 and minute virus of mice controls transcription. J. Virol. 64:387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutil, E. M., A. Toker, and A. C. Newton. 1998. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1). Curr. Biol. 8:1366-1375. [DOI] [PubMed] [Google Scholar]
- 24.Esdar, C., S. A. Oehrlein, S. Reinhardt, A. Maelicke, and T. Herget. 1999. The protein kinase C (PKC) substrate GAP-43 is already expressed in neural precursor cells, colocalizes with PKCeta and binds calmodulin. Eur. J. Neurosci. 11:503-516. [DOI] [PubMed] [Google Scholar]
- 25.Fima, E., M. Shtutman, P. Libros, A. Missel, G. Shafaf, G. Kahana, and E. Livneh. 2001. PKCeta enhances cell cycle progression, the expression of G1 cyclins and p21 in MCF-7 cells. Oncogene 20:6794-6804. [DOI] [PubMed] [Google Scholar]
- 26.Greif, H., J. Ben-Chaim, T. Shimon, E. Bechor, H. Eldar, and E. Livneh. 1992. The protein kinase C-related PKC-L(eta) gene product is localized in the cell nucleus. Mol. Cell. Biol. 12:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrington, E. O., K. E. Doyle, J. L. Brunelle, and J. A. Ware. 2000. Endothelial proliferation, migration, and differentiation are blunted by conditionally expressed protein kinase C pseudosubstrate peptides. Bioch. Biophys. Res. Commun. 271:499-508. [DOI] [PubMed] [Google Scholar]
- 28.Harris, C. E., R. A. Boden, and C. R. Astell. 1999. A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J. Virol. 73:72-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu, K. Q., J. M. Backer, G. Shagian, E. P. Feener, and G. L. King. 1990. Modulation of the insulin growth factorII/mannose 6-phosphate receptor in microvascular endothelial cells by phorbol ester via protein kinase C. J. Biol. Chem. 265:13864-13870. [PubMed] [Google Scholar]
- 30.Jaken, S., and P. J. Parker. 2000. Protein kinase C binding partners. Bioessays 22:245-254. [DOI] [PubMed] [Google Scholar]
- 31.Kashiwagi, M., M. Ohba, H. Watanabe, K. Ishino, K. Kasahara, Y. Sanai, Y. Taya, and T. Kuroki. 2000. PKCeta associates with cyclin E/cdk2/p21 complex, phosphorylates p21 and inhibits cdk2 kinase in keratinocytes. Oncogene 19:6334-6341. [DOI] [PubMed] [Google Scholar]
- 32.Kashiwagi, M., M. Ohba, K. Chida, and T. Kuroki. 2002. Protein kinase C eta (PKCeta): its involvement in keratinocyte differentiation. J. Biochem. 132:853-857. [DOI] [PubMed] [Google Scholar]
- 33.Kestler, J., B. Neeb, S. Struyf, J. van Damme, S. F. Cotmore, A. D'Abramo, P. Tattersall, J. Rommelaere, C. Dinsart, and J. J. Cornelis. 1999. Cis-requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum. Gene Ther. 10:1619-1632. [DOI] [PubMed] [Google Scholar]
- 34.Kornberg, A., and T. Baker. 1992. DNA replication., 2nd ed. W. H. Freeman & Co., New York, N.Y.
- 35.Krady, J. K., and D. C. Ward. 1995. Transcriptional activation by the parvoviral nonstructural protein NS-1 is mediated via a direct interaction with Sp1. Mol. Cell. Biol. 15:524-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legendre, D., and J. Rommelaere. 1992. Terminal regions of the NS1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J. Virol. 66:5705-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livneh, E., T. Shimon, E. Bechor, Y. Doki, I. Schieren, and I. B. Weinstein. 1996. Linking protein kinase C to the cell cycle: ectopic expression of PKC eta in NIH3T3 cells alters the expression of cyclins and Cdk inhibitors and induces adipogenesis. Oncogene 12:1545-1555. [PubMed] [Google Scholar]
- 38.Marks, F., and M. Gschwendt. 1996. Protein phosphorylation. VCH Verlagsgesellschaft mbH, Weinheim, Germany.
- 39.Newton, A. 1997. Regulation of protein kinase C. Curr. Opin. Cell Biol. 9:161-167. [DOI] [PubMed] [Google Scholar]
- 40.Nüesch, J. P. F., and P. Tattersall. 1993. Nuclear targeting of the parvoviral replicator protein molecule NS1: evidence for self-association prior to nuclear transport. Virology 196:637-651. [DOI] [PubMed] [Google Scholar]
- 41.Nüesch, J. P. F., S. F. Cotmore, and P. Tattersall. 1992. Expression of functional parvoviral NS1 from recombinant vaccinia virus: effects of mutations in the nucleotide-binding motif. Virology 191:406-416. [DOI] [PubMed] [Google Scholar]
- 42.Nüesch, J. P. F., J. Christensen, and J. Rommelaere. 2001. Initiation of minute virus of mice DNA replication is regulated at the level of origin unwinding by atypical protein kinase C phosphorylation of NS1. J. Virol. 75:5730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nüesch, J. P. F., R. Corbau, P. Tattersall, and J. Rommelaere. 1998. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated by the phosphorylation state of the polypeptide J. Virol. 72:8002-8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nüesch, J. P. F., S. F. Cotmore, and P. Tattersall. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209:122-135. [DOI] [PubMed] [Google Scholar]
- 45.Nüesch, J. P. F., S. Dettwiler, R. Corbau, and J. Rommelaere. 1998. Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J. Virol. 72:9966-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nüesch, J. P. F., S. Lachmann, R. Corbau, and J. Rommelaere. 2003. Regulation of minute virus of mice NS1 replicative functions by atypical PKCλ in vivo. J. Virol. 77:433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oehrlein, S. A., A. Maelicke, and T. Herget. 1998. Expression of protein kinase C gene family members is temporally and spatially regulated during neural development in vitro. Eur. J. Cell Biol. 77:323-337. [DOI] [PubMed] [Google Scholar]
- 48.Ohno, S. 2001. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr. Opin. Cell Biol. 13:641-648. [DOI] [PubMed] [Google Scholar]
- 49.Op de Beek, A., F. Anouja, S. Mousset, J. Rommelaere, and P. Caillet-Fauquet. 1995. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 6:781-787. [PubMed] [Google Scholar]
- 50.Osada, S., K. Mizuno, T. C. Saido, Y. Akita, K. Suzuki, T. Kuroki, and S. Ohno. 1990. A phorbol ester receptor/protein kinase, nPKC eta, a new member of the protein kinase C family predominantly expressed in lung and skin. J. Biol. Chem. 265:22434-22440. [PubMed] [Google Scholar]
- 51.Parker, J. S., W. J. Murphy, D. Wang, S. J. O'Brien, and C. R. Parrish. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pujol, A., L. Deleu, J. P. F. Nüesch, C. Cziepluch, J.-C. Jauniaux, and J. Rommelaere. 1997. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of the nonstructural protein NS1. J. Virol. 71:7393-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhode, S. L., III, and S. M. Richard. 1987. Characterization of the trans-activation-responsive element of the parvovirus H-1 P38 promoter. J. Virol. 61:2807-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rommelaere, J., and J. J. Cornelis. 1991. Antineoplastic activity of parvoviruses. J. Virol. Methods 33:233-251. [DOI] [PubMed] [Google Scholar]
- 55.Schonhorn, J. E., T. Akompong, and M. Wessling-Resnick. 1995. Mechanism of transferrin receptor down-regulation in K562 cells in response to protein kinase C activation. J. Biol. Chem. 270:3698-3705. [DOI] [PubMed] [Google Scholar]
- 56.Schönwasser, D., R. M. Marais, C. J. Marschall, and P. J. Parker. 1998. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol. Cell. Biol. 18:790-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shih, M., F. Lin, J. D. Scott, H. Y. Wang, and C. C. Malbon. 1999. Dynamic complexes of beta2-adrenergic receptors with protein kinases and phosphates and the role of gravin. J. Biol. Chem. 274:1588-1595. [DOI] [PubMed] [Google Scholar]
- 58.Tobaben, S., P. Thakur, R. Fernandez-Chacon, T. C. Sudhof, J. Rettig, and B. Stahl. 2001. A trimeric protein complex functions as a synaptic chaperone machine. Neuron 31:987-999. [DOI] [PubMed] [Google Scholar]
- 59.Toker, A. 1998. Signaling through protein kinase C. Front. Biosci. 3:D1134-D1147. [DOI] [PubMed] [Google Scholar]
- 60.Vanacker, J.-M., R. Corbau, G. Adelmant, M. Perros, V. Laudet, and J. Rommelaere. 1996. Transactivation of a cellular promoter by the NS1 protein of the parvovirus minute virus of mice through a putative hormone-responsive element. J. Virol. 70:2369-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veronese, S., M. Gambacorta, and B. Falini. 1989. In situ demonstration of tissue proliferative activity using anti-bromo-deoxyuridine monoclonal antibody. J. Clin. Pathol. 42:820-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson, G. M., H. K. Jindal, D. E. Yeung, W. Chen, and C. R. Astell. 1991. Expression of minute virus of mice major nonstructural protein in insect cells: purification and identification of ATPase and helicase activities. Virology 185:90-98. [DOI] [PubMed] [Google Scholar]