Abstract
The transmembrane protein of Mason-Pfizer monkey virus contains two heptad repeats that are predicted to form amphipathic α-helices that mediate the conformational change necessary for membrane fusion. To analyze the relative sensitivity of the predicted hydrophobic face of the N-terminal heptad repeat to the insertion of uncharged, polar, and charged substitutions, mutations that introduced alanine, serine, or glutamic acid into positions 436, 443, 450, and 457 of the envelope protein were examined. Novel systems using Tat protein and the GHOST cell line were developed to test and quantitate the effects of the mutations on Env-mediated fusion and infectivity of the virus. While no single amino acid change at any of the positions interfered significantly with the synthesis, processing, or transport to the plasma membrane of glycoprotein complexes, 9 of the 12 nonconservative mutations in these residues completely abolished fusion activity and virus infectivity. Mutations in the central positions (443 and 450) of the heptad repeat region were the most detrimental to Env function, and even single alanine substitutions in these positions dramatically altered the fusogenicity of the protein. These results demonstrate that this N-terminal heptad repeat plays a critical role in Env-mediated membrane fusion and highlight the key function of central hydrophobic residues in this process and the sensitivity of all positions to charge substitutions.
Mason-Pfizer monkey virus (M-PMV) was the first Betaretrovirus to be isolated from a nonhuman primate and was recovered from a spontaneous breast carcinoma of a rhesus monkey (13, 29). Subsequent studies revealed that this virus, although isolated from a mammary adenocarcinoma, was not oncogenic (19); instead, infected macaques suffered a severe immunodeficiency syndrome with pathology distinct from those of lentiviruses, such as simian immunodeficiency virus and human immunodeficiency virus (HIV) (14). Betaretroviruses are characterized by the assembly of intracytoplasmic capsids, which make their way to the plasma membrane and are released by budding. Most retroviruses, in contrast, assemble their capsids and bud simultaneously from the plasma membrane.
Retrovirus glycoproteins are translated as a polyprotein precursor from a spliced env gene-specific mRNA. The glycosylated precursor is assembled into oligomers in the endoplasmic reticulum and then proteolytically cleaved by a host protease into two subunits, the surface (SU) and transmembrane (TM) proteins, in the late Golgi complex (28). These glycoprotein complexes are then incorporated into budding virions at the plasma membrane. The SU glycoprotein is responsible for cellular tropism for the virus, whereas the TM glycoprotein is responsible for anchoring the SU protein in the viral membrane and for mediating virus-cell membrane fusion during viral entry. The glycoprotein precursor of M-PMV, Pr86, is cleaved to yield the mature gp70 (SU) and gp22 (TM) proteins, and then, following virus release, a viral protease-mediated maturational cleavage of the gp22 cytoplasmic domain results in conversion of gp22 into gp20 (2-4). The TM glycoprotein of M-PMV, like those of other retroviruses, can be divided into three regions—the extracellular domain, the membrane-spanning domain, and the cytoplasmic domain (28). Within the extracellular domain, there are two regions that exhibit the characteristics of heptad repeats (HRs).
HR motifs are characterized by the ability to form amphipathic helices (1, 33). A specialized form, the leucine zipper, contains repeats of leucine at every seventh position (the first of these, by convention, is termed the “a” position) (32, 36). In addition, there tend to be nonpolar amino acids at the fourth, or “d,” position within the α-helix. These helices specifically associate along a hydrophobic interface to form a coiled-coil structure (25, 26, 35). The chemical nature of the hydrophobic residues in the a and d positions can influence the valency of interactions so that trimers and tetramers can assemble (27).
HR sequences have also been detected in the TM proteins of the paramyxoviruses (5, 8, 31, 37, 43), influenza virus (42, 45, 47), coronavirus (8, 15, 34; P. Britton, Letter, Nature 353:394, 1991), and other retroviruses (8, 17, 21, 22) and have been shown to play an essential role in viral fusion and infectivity (12, 17, 23, 34, 41, 44, 46). X-ray crystallographic studies of the influenza virus hemagglutinin subunit 2 (HA2), the TM subunit of Moloney murine leukemia virus, and the HIV Env TM subunit demonstrated that the HR regions formed coiled-coil trimer structures (6, 9, 18). In M-PMV, the N-terminal HR domain (HR1) is located in the extracellular domain of gp22 (residues 436 to 457). To analyze the relative sensitivity of the predicted hydrophobic face of HR1 to the insertion of uncharged, polar, and charged substitutions, mutations that introduced alanine, serine, or glutamic acid into positions 436, 443, 450, and 457 of the envelope (Env) protein were examined. Novel systems using the Tat protein and the GHOST cell line were developed to test and quantitate the effects of the mutations on the Env-mediated fusion and infectivity of the virus. These studies indicate that HR1 plays a critical role in M-PMV membrane fusion and virus entry and that the central a positions in this domain are the most critical for this function.
MATERIALS AND METHODS
Cells, culture, and transfections.
African green monkey kidney (COS-1) cells were obtained from the American Type Culture Collection. The HOS-CD4/LTR-hGFP (GHOST) cell line, which expresses green fluorescent protein (GFP) under the control of an HIV long terminal repeat (LTR), was obtained though the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, and was originally contributed by Vineet N. KewalRamani and Dan R. Littman. The cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum. The GHOST cells were additionally maintained in medium containing hygromycin and geneticin (G418) as recommended by the contributor. Each mutant DNA was transfected into COS-1 cells by a modified calcium phosphate technique (11).
Construction of plasmids and site-directed mutagenesis.
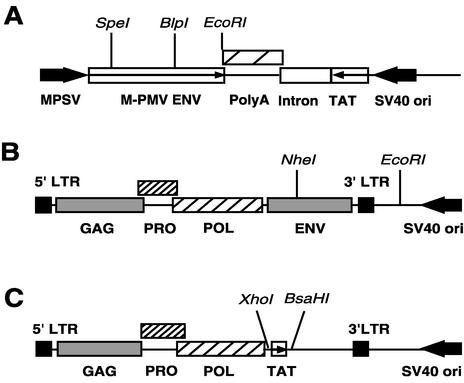
The pTMT vector is an M-PMV glycoprotein expression vector based on the plasmid pTMO (3) in which the Neor gene was replaced by the HIV type 1 (HIV-1) tat gene. The tat gene, under the transcriptional control of the simian virus 40 (SV40) early promoter and containing an SV40-derived poly(A) sequence, was removed from pSV2tat72 (20), which was obtained from the NIH AIDS Research and Reference Reagent Program, by digestion with PvuII and EcoRI. The pTMO vector containing the M-PMV glycoprotein gene under the transcriptional control of the myeloproliferative sarcoma virus promoter was linearized by digestion with BamHI. This fragment was blunt ended with the Klenow fragment of DNA polymerase I and then digested with EcoRI to remove the Neor gene. The 1.8-kb fragment from pSV2tat72 was ligated to the 6.1-kb vector fragment from pTMO, and the resulting plasmid was named pTMT (Fig. 1A).
FIG. 1.
Schematic representation of the M-PMV constructs. (A) Schematic diagram of the M-PMV glycoprotein expression vector, pTMT, in which the env gene is under the control of the myeloproliferative sarcoma virus promoter and the tat gene is under the control of the SV40 early promoter. Large arrows show the direction of transcription from the MPSV and SV40 promoters. Small arrows show the direction in which protein is translated. (B) Schematic diagram of pSARM4, an M-PMV expression vector. The plasmid pSARM4 contains an infectious M-PMV genome. (C) Schematic diagram of pMTΔE vector. The plasmid pMTΔE is a replication-defective M-PMV genome in which the env gene has been replaced by coding sequences for the HIV-1 tat gene.
pSARM4 (Fig. 1B), which is a plasmid containing the entire infectious M-PMV proviral genome, was constructed in the following manner. First the pSHRM15 plasmid (38) was digested with AvaI to remove the hygromycin gene. The vector fragment was self-ligated, and then an XbaI site at the 5′ boundary of the flanking cellular sequences was changed into an XmaI site by PCR mutagenesis, and a new XmaI site was introduced adjacent to the 5′ LTR of the M-PMV genome. The plasmid was then digested with XmaI and blunt ended to remove the flanking sequences from the plasmid. The resulting vector fragment was self-ligated to generate the pSARM4 plasmid, which exhibits enhanced stability compared to pSHRM15 when propagated in Escherichia coli.
The pMTΔE vector (Fig. 1C) is a replication-defective M-PMV genome in which the env gene has been replaced by coding sequences for the HIV-1 tat gene. For this construction, we introduced an XhoI site into nucleotide 5185 of pSARM4 to yield pSARMX. The latter plasmid was digested with XhoI and BasHI, and the env− fragment was ligated to an XhoI-BasHI fragment from pSV2tat72 that contains tat.
In order to change each “a” position of the HR into alanine, serine, or glutamic acid, site-directed mutagenesis was carried out using the “mega primer” PCR method. A small fragment of the EcoRI-digested pTMT vector was purified and then used as a template for PCR mutagenesis. In the first PCR step, a reverse primer (5′-CTGGGCGCATGCTCAGTAGC) which could anneal to a sequence in the gp22 coding region was designed and used, in combination with mutagenic primers for each corresponding site, to amplify the DNA sequence. The sequences of the mutagenic oligonucleotides used were as follows: V436A, 5′-ATATCAGATGCTCAAGCTATT; V436S, 5′-ATATCAGATAGTCAAGCTATT; V436E, 5′-ATATCAGATGAACAAGCTATT; I443A, 5′-TCTAGCACTGCACAAGATCTC; I443S, 5′-TCTAGCACTAGTCAAGATCTC; I443E, 5′-TCTAGCACTGAACAAGATCTC; V450A, 5′-CAAGATCAGGCAGACTCTCTA; V450S, 5′-CAAGATCAGTCAGACTCTCTA; V450E, 5′-CAAGATCAGGAAGACTATCT; V457A, 5′-CAGAAGTAGCACTGCAAAAC; V457S, CAGAAGTATGACTGCAAAAC; and V457E, 5′-GCAGAAGTAGAACTGCAAAAC.
After the first-step PCR, all the PCR products were purified. For the second-step PCR, a primer (5′-AGGTCTAGCTAGCTTTACTA) which could anneal to a sequence in the gp22 coding region was designed and used in combination with the first PCR products as a reverse primer. After the second-step PCR, the products were digested with BlpI and SpeI and cloned into the same unique sites in pTMT. The mutated sequences were excised from the pTMT vectors by digestion with EcoRI and NheI, and the resulting fragment was inserted into the proviral vector pSARM4, which has unique EcoRI and NheI sites. All of the mutations were confirmed by DNA sequencing.
Protein expression, radioactive labeling, and immunoprecipitation.
The env expression plasmid pTMT was transfected into COS-1 cells in 100-mm-diameter plates by a modified CaPO4 technique for the analysis of glycoprotein expression. At 36 h posttransfection, the cells were divided into two sets of 60-mm-diameter plates, one for pulse-labeling and the other for pulse-chase labeling. At 72 h posttransfection, the cells were starved in leucine-free DMEM for 90 min and then pulse-labeled in leucine-free DMEM supplemented with [3H]leucine (1 mCi/ml; 0.25 ml/plate) for 30 min. At this point, one set of pulse-labeled cells was lysed with lysis buffer A (1% Triton X-100, 15% sodium deoxycholate, 0.15 M NaCl, 0.05 M Tris, pH 7.5) on ice. The other set was then chased in complete DMEM for an additional 4 h prior to lysis of the cells. Using a goat anti-M-PMV antibody, viral proteins in the cell lysates were immunoprecipitated and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (39).
Fusion assays of wild-type and mutant M-PMV Env proteins.
The fusion activity of the M-PMV Env protein was measured by a combination of GFP expression and flow cytometric analysis. Fusion of COS-1 cells transfected with the pTMT vector, which expresses both M-PMV Env and HIV-1 Tat, with GHOST cells resulted in expression of GFP. These GFP-positive fused cells were then quantitated by flow cytometery. The glycoprotein expression vector pTMT, containing either wild-type or mutant M-PMV env genes, was transfected into COS-1 cells by a modified CaPO4 method, and 36 h posttransfection, the cells were trypsinized, mixed in a 1:2 ratio with untransfected GHOST indicator cells, and replated in a six-well plate. After 24 to 30 h of incubation, the cells were washed twice with deficient phosphate-buffered saline (PBS) (PBS without 0.1 mM CaCl2 and 1 mM MgCl2), and then 500 μl of PBS-1 mM EDTA was added to resuspend the cells. The cells were kept on ice at all times. The resuspended cells were analyzed for GFP expression using a FACScan flow cytometer (Becton Dickinson) equipped with a 488-nm argon laser. Gating on the living cells was achieved by the use of light-scattering measurements. Using the parameters of forward scatter and side scatter in a dot plot, it was possible to estimate cell size and granularity. Since M-PMV Env proteins generally yield syncytia containing only two or three nuclei, we selected cells that were larger than the single cell population to maximize the efficiency of the assay. The histogram of the fluorescence intensities of the living cells was further used to distinguish between the cells fused by the M-PMV Env proteins and the larger unfused-cell populations.
Viral protein expression and glycoprotein incorporation.
COS-1 cells, in 100-mm-diameter plates, were transfected with 10 μg of the molecular clone pSARM4, containing either the wild-type or mutant env gene, using the modified CaPO4 method. Three days after transfection, the cells were labeled with 500 μCi of [3H]leucine in 0.8 ml of leucine-deficient DMEM. The cells were labeled for 30 min, at which time the label was removed, complete DMEM containing 10% fetal bovine serum was added, and incubation was continued for 6 h. The supernatant was collected, and following removal of the cells debris by filtration though a 0.45-μm-pore-size filter, the supernatant was loaded onto a 25% (wt/vol) sucrose cushion in PBS and centrifuged for 30 min in a TLA 100.3 rotor (Beckman) at 100,000 rpm. The pellet was resuspended in lysis buffer B, and M-PMV viral proteins were immunoprecipitated as described above with goat anti-M-PMV antiserum. The immunoprecipitates were analyzed by SDS-12% PAGE and fluorography. In some instances, for analysis of gp20/gp22 incorporation into virions, the pelleted virions were analyzed directly by SDS-PAGE.
Single-round virus infectivity assay.
COS-1 cells in 100-mm-diameter plates were cotransfected with both the glycoprotein expression vector pTMT, containing either the wild-type or the mutant env gene, and pMTΔE, which carries the tat gene of HIV-1 in place of env, by using a modified CaPO4 method. At 48 h after cotransfection, culture supernatants were collected and filtered through a 0.45-μm-pore-size filter. Relative levels of reverse transcriptase (RT) activity were determined for each sample as previously described (10), and the RT levels were normalized by dilution with complete medium. The normalized supernatants were used to infect GHOST cells in duplicate. After 48 h of incubation, the cells were washed twice with deficient PBS, and 500 μl of PBS- 1 mM EDTA was added to resuspend the cells. The resuspended cells were analyzed for GFP expression on a flow cytometer as described above for the fusion assay, except all living cell populations were selected instead of just those larger than single cells.
RESULTS
Mutagenesis of the N-terminal HR region of the M-PMV transmembrane glycoprotein.
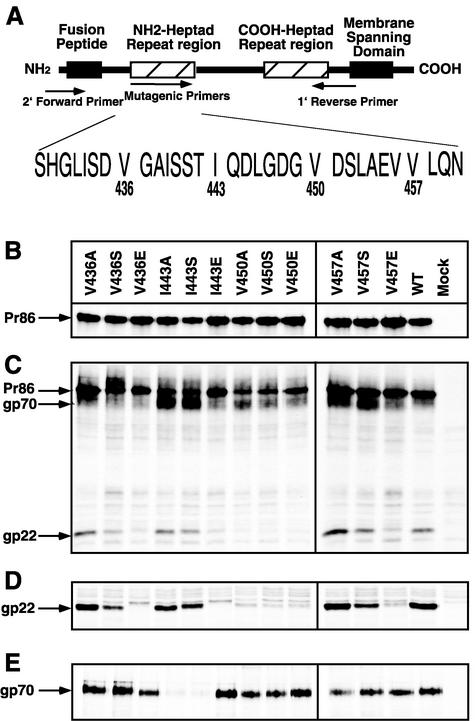
The HR1 domain of the M-PMV TM is located in the ectodomain of gp22 and contains repeats of either valine or isoleucine at every seventh position (Fig. 2A). In addition, leucine or isoleucine residues are located at the fourth, or “d,” position within the HR. A series of site-directed mutations that change the “a” positions of the HR region into alanine, serine, or glutamic acids were introduced by PCR mutagenesis. The nomenclature for the mutants followed the convention of a letter designation for a wild-type amino acid, followed by the residue number and the substituted amino acid (e.g., V436A). The mutant genes were cloned into two different expression vectors, pTMT and pSARM4. All mutations were confirmed through DNA sequencing to ensure that no additional mutations had been introduced.
FIG. 2.
Synthesis and processing of mutant and wild-type (WT) glycoproteins. (A) Diagram of the M-PMV TM protein showing the location of the HR1 domain of M-PMV relative to the rest of the glycoprotein. The positions of the primers used to introduce mutations are shown. (B) COS-1 cells were transfected with the pTMT expression vector containing either wild-type or mutant env genes. At 48 h posttransfection, the cells were labeled with [3H]leucine for 30 min and immunoprecipitated with goat anti-M-PMV serum as described in the text. The mutant designations are shown above each lane, and the position of the precursor glycoprotein, Pr86, is indicated to the left. Mock, mock transfected. (C) Processing of the env gene polyprotein precursor protein. Following a 4-h chase in unlabeled medium, Env proteins were immunoprecipitated with goat anti-M-PMV serum and electrophoresed in SDS-PAGE. The positions of the precursor glycoprotein, Pr86, and the cleavage products, gp70 and gp22, are shown. (D) Position of gp22 after longer exposure. (E) Secretion of SU into culture media. Following pulse-chase, the culture media were collected, immunoprecipitated with goat anti-M-PMV serum, and analyzed for the released SU proteins.
Expression of mutant envelope glycoproteins.
In order to determine the effects of the mutations on Env expression, processing, and transport, COS-1 cells were transfected with each of the pTMT vectors containing either mutant or wild-type env genes. At 72 h posttransfection, the proteins were metabolically labeled in a pulse-chase experiment, and the resulting products were immunoprecipitated from both the cell lysates and the cell culture medium with goat anti-M-PMV antiserum. The immunoprecipitates were analyzed on SDS-PAGE and subjected to autoradiography (Fig. 2).
All of the mutant glycoprotein precursor products were expressed at levels similar to that of the wild-type protein (Fig. 2B), and all were processed into gp70 and gp22 (Fig. 2C). However, we consistently observed accumulation of a higher-molecular-weight band in mutant V436S-transfected cells, suggesting slower processing of this mutant protein. The gp22 proteins of six mutants, V436E, I443E, V450A, V450S, V450E, and V457E, were immunoprecipitated less efficiently (Fig. 2D), even though the levels of cell-associated gp70 were equivalent to those of the wild type, suggesting that these mutations interfere with anti-TM antibody binding to the mutant gp22 protein. Because some of the noncovalently linked gp70 of M-PMV is secreted into the culture medium (2, 3), we utilized this as an independent measure of Env transport to the plasma membrane. With the exception of I443A and I443S, similar amounts of secreted gp70 were immunoprecipitated from culture media collected after a 4-h pulse-chase of COS-1 cells transfected with either mutant or wild-type pTMT (Fig. 2E). For these two mutants, reduced amounts of gp70 were secreted into the culture medium and increased levels of gp70 were observed in the cell lysates (Fig. 2C).
Viral protein expression and glycoprotein incorporation into budding virus.
To test if the mutations introduced into the N-terminal HR affected the incorporation of M-PMV glycoprotein into virions, the mutant env genes were cloned into the infectious molecular clone pSARM4. Each clone, containing either the wild-type or mutated env gene, was transfected into COS-1 cells, and 72 h posttransfection, the cells were labeled with [3H]leucine for 30 min. Following a 6-h chase in complete medium, virus particles in the supernatant were filtered through a 0.45-μm-pore-size filter, pelleted through a 25% sucrose cushion, and then immunoprecipitated as described in Materials and Methods.
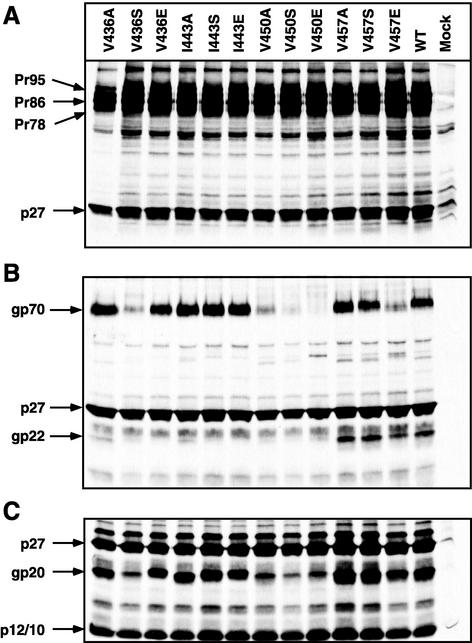
The pattern of cell-associated viral Env and Gag proteins expressed from wild-type pSARM4 is shown in Fig. 3A, lane WT. After the pulse-chase labeling, the viral gag gene products, Pr95, Pr78, and p27, and the Env precursor, Pr86, could be immunoprecipitated. A similar pattern of viral proteins was observed when the mutant proviral constructs were analyzed. The patterns of viral Env and Gag proteins immunoprecipitated from virus particles pelleted through a 25% sucrose cushion are shown in Fig. 3B. For the wild-type virions, the mature env gene products gp70 and gp20, as well as the gag gene products p27 and p10/p12, could be immunoprecipitated with goat anti-M-PMV antibody (3). Five mutants (V436S, V450A/S/E, and V457E) showed reduced or no gp70 incorporation, while the remaining mutants incorporated levels of gp70 equivalent to that of the wild type. However, for a majority of the mutants, a significant discrepancy was observed between the levels of virus-associated gp70 and gp20 (Fig. 3B). These results paralleled those from the pTMT vector (Fig. 2C) and suggested that the mutations interfere with antibody binding to a critical conformational epitope in gp20.
FIG. 3.
Expression of envelope glycoprotein mutants in the context of provirus. The mutant env genes in pSARM4 were transfected into COS-1 cells and metabolically labeled as described in Materials and Methods. (A) Cell lysates were immunoprecipitated with goat anti-M-PMV serum and analyzed in SDS-PAGE. The mutant designation is shown above each lane, and the positions of the viral bands are indicated on the left. WT, wild type; Mock, mock transfected. (B and C) Incorporation of mutant glycoprotein into virions. Virus-containing supernatants from metabolically labeled COS-1 cells transfected with either wild-type or mutant pSARM4 constructs were centrifuged through a 25% sucrose cushion. The virus pellets were recovered and immunoprecipitated with goat anti-M-PMV serum (B) or electrophoresed directly (C). The positions of the viral bands are indicated on the left.
To test this hypothesis, the viral pellet from the sucrose cushion was resuspended directly in protein-loading buffer and analyzed by SDS-PAGE. Mutants V436A/E, I443A/S/E, and V457A/S had similar or somewhat higher levels of gp20 proteins than the wild type (Fig. 3C), indicating that the mutations at these sites have no effect on the incorporation of glycoprotein into virions. Even though mutants V436S, V450A/S/E, and V457E were transported to the plasma membrane and secreted gp70 at wild-type levels, substitutions in these sites reduced the incorporation of the proteins into virions (Fig. 3B). These mutants were therefore not analyzed further.
Effects of the mutations on cell-cell fusion.
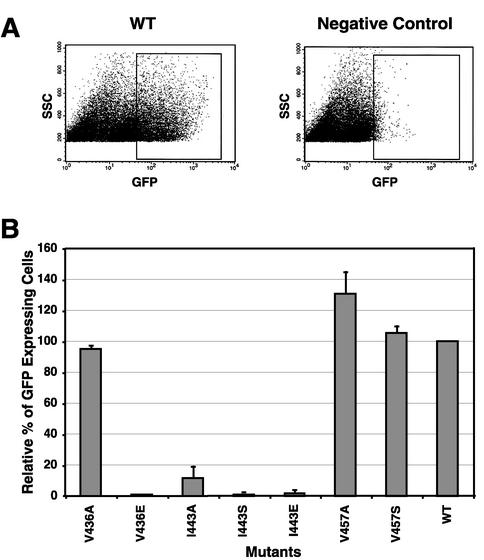
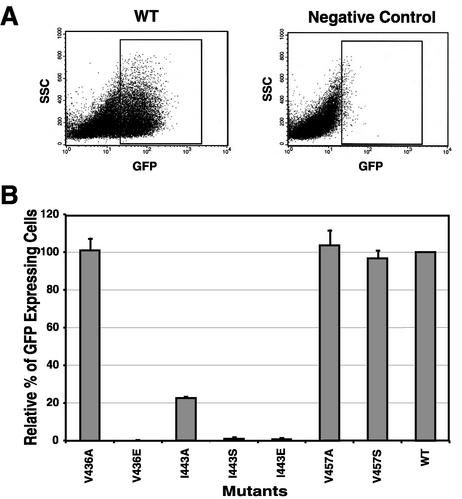
M-PMV Env is a poorly fusogenic protein compared to that of HIV-1. Wild-type Env-induced syncytia (two to four nuclei) are difficult to distinguish from background multinucleated cells (two or three nuclei) (4). In order to quantitatively assess the fusion activities of different M-PMV Env proteins, we have developed a novel, highly sensitive method that utilizes GFP and flow cytometry. Determination of the average number of GFP-expressing cells using fluorescence-activated cell sorter (FACS) analysis typically provides a more consistent and reproducible measure of this biological activity than the determination of the average number of nuclei per syncytium or the number of syncytia per plate. Briefly, COS-1 cells were transfected with each of the env expression pTMT vectors containing either mutant env genes or wild-type env. At 36 h posttransfection, the transfected cells were mixed with GHOST cells at a ratio of 1:2. After another 24 h of incubation, cells were removed from the plate and analyzed on a FACScan flow cytometer (Fig. 4A). For the negative controls, COS-1 cells were transfected with the pTMTΔEnv vector, which can express Tat protein in the transfected cells (40). Relative levels of fusion were calculated for each mutant compared to wild-type Env, and the results of three independent experiments are summarized in Fig. 4B.
FIG. 4.
Cell-cell fusion assay of envelope glycoproteins expressed from the pTMT expression vector. COS-1 cells were transfected with the pTMT expression vector containing either the mutant or wild-type (WT) env gene. COS-1 cells expressing glycoproteins were mixed 1:2 with GHOST cells, replated, and analyzed as described in Materials and Methods. (A) Cells were gated on size and then analyzed for expression of GFP by quantitating excitation at 488 nm. (B) The percentage of fused GFP-positive cells was calculated for each mutant and normalized to the wild type as shown. The graph depicts the mean of three independent experiments (± standard deviation). The mutant designation is shown below each histogram.
The mutants V436A, V457A, and V457S generated similar or somewhat higher numbers of GFP-expressing cells than the wild type, indicating that these three mutations had little effect on the fusogenicity of the TM protein. In contrast, any substitution, including alanine, in the central position (residue 443) of the HR region dramatically reduced fusion activity (Fig. 4B).
Effects of the mutations on single-round virus infectivity.
In order to analyze the effects of the mutations introduced into the N-terminal HR on virus infectivity, a complementation assay was used. In this single-cycle assay, HR1 mutants that incorporated normal levels of Env were assayed for the ability to complement a vector (pMTΔE) that carries the tat gene of HIV-1 in place of the M-PMV env gene. The pTMT and pMTΔE vectors were cotransfected into COS-1 cells, and 2 days after transfection, the culture supernatants were assayed for RT activity as described previously (10). Equivalent amounts of RT-containing supernatant were used to infect GHOST cells, and 2 days postinfection, the cells were analyzed by flow cytometry for the number of GFP-positive cells (Fig. 5A). As negative controls, the assay was also performed by transfecting COS-1 cells with either the pMTΔE vector or the pTMT vector alone, to measure the background GFP expression level in the GHOST cells (Fig. 5A).
FIG. 5.
Single-round infectivity assay of virus. COS-1 cells were cotransfected with pTMT and pMΤΔΕ expression vectors as described in Materials and Methods. Culture medium from cells expressing virus was filtered and normalized for RT activity. The normalized medium was used to infect GHOST cells. (A) Cells were gated on size and granularity and then analyzed for GFP expression. WT, wild type. (B) A total of 3 × 104 cells were analyzed for each construct, and the number of GFP-expressing cells relative to the wild type was plotted. The graph shows the mean from three independent experiments (± standard deviation).
Three mutants, V436A, V457A, and V457S, showed levels of infectivity equivalent to that of the wild type. Mutant V443A, which showed a reduced level of activity in the cell-cell fusion assay, had a similarly low level of infectivity, while mutants V436E and V443S/E were essentially noninfectious (Fig. 5B) despite incorporating normal levels of Env (Fig. 3B). The results of this infectivity assay thus paralleled those observed from the cell-cell fusion assay.
DISCUSSION
Structural studies of the TM protein of HIV-1, simian immunodeficiency virus, and murine leukemia virus have shown that HR regions located in the N terminus (HR1) and C terminus (HR2) of the ectodomain can form a coiled-coil six-helix structure. Based on crystallographic information from the influenza virus HA2 trimer, the formation of this six-helix structure is hypothesized to be critical for membrane fusion, first by relocating the fusion peptide into the target cell membrane by formation of an N-terminal HR coiled-coil structure and second by mechanically bringing the membranes together through packing of C-terminal HR domains into a hydrophobic groove in the coiled coil (7). Genetic evidence supporting this model of retrovirus fusion has been obtained for HIV-1, in which mutations that prevent coiled-coil formation by the N-terminal HR abrogate fusion (17, 46). The experiments described here were designed to investigate the effect of engineering neutral, polar, or acidic amino acid substitutions into the hydrophobic “a” positions of the similarly located HR1 of M-PMV. In order to quantitatively assess the biological consequences of those substitutions, it was necessary to develop novel assays for M-PMV Env-mediated cell-cell fusion and virus infectivity. These methodologies, which employ HIV-1 Tat-induced activation of GFP, have proven to be robust and highly reproducible and may be applicable to other glycoprotein-mediated fusion systems.
Analysis of cell-cell fusion mediated by the mutated env genes showed that the two central a positions of the N-terminal HR domain are particularly sensitive to amino acid substitutions; all three of the I443 mutants which were expressed normally on the cell surface and incorporated into virions had dramatically reduced ability to mediate fusion. Similar results were obtained with each of the V450 mutants (data not shown). In contrast, the flanking a positions (V436 and V457) were relatively insensitive to the substitution of a neutral alanine residue. Analyses of alanine substitution mutants in the leucine zipper domain of the paramyxovirus F protein (37) and coronavirus S protein (34) have revealed similar results in that leucine residues at the ends of the HR domain are less sensitive than those in the middle.
In general, it appears that the more polar the substitution, the more impaired fusogenicity is for each of the mutant N-terminal HRs. This is consistent with our previous observations with the analogous repeat of HIV-1, where substitution of alanine, serine, or proline into a highly conserved isoleucine residue (I573) results in a progressive decrease in the melting temperature of the coiled coil and in the fusogenicity of Env (17, 46). The single-round infectivity assay showed that the three substitution mutants (V436A, V457A, and V457S) that possess wild-type fusion activity can also mediate virus entry at wild-type levels, while with the exception of I443A, which retained 20% infectivity, all of the mutants that exhibit reduced levels of cell fusion display almost background levels of infectivity.
Four of the mutant viruses (V436E, I443A, I443S, and I443E) had levels of SU equivalent to that of the wild type in virus but had undetectable or low levels of gp20 in immunoprecipitates. This did not reflect the absence of the TM protein from virions, since direct analysis of pelleted virus revealed wild-type levels of TM (Fig. 3C). This indicated that these mutations abrogate the binding of antibodies to an immunodominant epitope in the protein. A similar effect was observed previously when a small deletion was introduced into the TM protein C terminal of the HR (3). Several antibodies have recently been described that bind to the six-helix bundle formed from HR1 and HR2 (16, 24, 30). This structure has been shown to form spontaneously following detergent solubilization of the Env complex from mammalian cell membranes (16). Thus, it is possible that the loss of antibody binding observed with these diverse mutations, which abrogate biological activity, also reflects the blocking of six-helix bundle formation. Such a possibility would also be consistent with the normal transport and incorporation of these mutants. Mutations in HR2 that also block six-helix bundle formation should allow us to test this hypothesis.
In summary, this study of the HR1 domain in the TM protein of M-PMV has demonstrated that the amino acids in the central “a” positions (I443 and V450) are indispensable for the formation of a fusion-active TM protein. Even though the amino acid residues at the ends of the HR domain are less sensitive to changes, the introduction of acidic residues into the same sites abrogates the biological function of the Env protein.
Acknowledgments
We thank Susan Dubay and Tshana Thomas for excellent technical assistance.
This work was supported by grant CA-27834 from the National Institutes of Health to E.H. FACS analysis was performed at the Flow Cytometry Core Facilities of the University of Alabama at Birmingham Center for AIDS Research (supported by NIH grant P30-AI-27767).
REFERENCES
- 1.Alber, T. 1992. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 2:205-210. [DOI] [PubMed] [Google Scholar]
- 2.Brody, B. A., and E. Hunter. 1992. Mutations within the env gene of Mason-Pfizer monkey virus: effects on protein transport and SU-TM association. J. Virol. 66:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brody, B. A., M. G. Kimball, and E. Hunter. 1994. Mutations within the transmembrane glycoprotein of Mason-Pfizer monkey virus: loss of SU-TM association and effects on infectivity. Virology 202:673-683. [DOI] [PubMed] [Google Scholar]
- 4.Brody, B. A., S. S. Rhee, and E. Hunter. 1994. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J. Virol. 68:4620-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckland, R., E. Malvoisin, P. Beauverger, and F. Wild. 1992. A leucine zipper structure present in the measles virus fusion protein is not required for its tetramerization but is essential for fusion. J. Gen. Virol. 73:1703-1707. [DOI] [PubMed] [Google Scholar]
- 6.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 7.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, P., C. R. Pringle, and A. J. Easton. 1990. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 71:3075-3080. [DOI] [PubMed] [Google Scholar]
- 9.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee, S., J. Bradac, and E. Hunter. 1985. A rapid screening procedure for the isolation of nonconditional replication mutants of Mason-Pfizer monkey virus: identification of a mutant defective in pol. Virology 141:65-76. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, S. S., S. F. Lee, H. J. Hao, and C. K. Chuang. 1998. Mutations in the leucine zipper-like heptad repeat sequence of human immunodeficiency virus type 1 gp41 dominantly interfere with wild-type virus infectivity. J. Virol. 72:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chopra, H. C., and M. M. Mason. 1970. A new virus in a spontaneous mammary tumor of a rhesus monkey. Cancer Res. 30:2081-2086. [PubMed] [Google Scholar]
- 14.Daniel, M. D., N. W. King, N. L. Letvin, R. D. Hunt, P. K. Sehgal, and R. C. Desrosiers. 1984. A new type D retrovirus isolated from macaques with an immunodeficiency syndrome. Science 223:602-605. [DOI] [PubMed] [Google Scholar]
- 15.de Groot, R. J., W. Luytjes, M. C. Horzinek, B. A. van der Zeijst, W. J. Spaan, and J. A. Lenstra. 1987. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 196:963-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Rosny, E., R. Vassell, P. T. Wingfield, C. T. Wild, and C. D. Weiss. 2001. Peptides corresponding to the heptad repeat motifs in the transmembrane protein (gp41) of human immunodeficiency virus type 1 elicit antibodies to receptor-activated conformations of the envelope glycoprotein. J. Virol. 75:8859-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubay, J. W., S. J. Roberts, B. Brody, and E. Hunter. 1992. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J. Virol. 66:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 19.Fine, D. L., J. C. Landon, R. J. Pienta, M. T. Kubicek, M. G. Valerio, W. F. Loeb, and H. C. Chopra. 1975. Responses of infant rhesus monkeys to inoculation with Mason-Pfizer monkey virus materials. J. Natl. Cancer Inst. 54:651-658. [PubMed] [Google Scholar]
- 20.Frankel, A. D., and C. O. Pabo. 1988. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55:1189-1193. [DOI] [PubMed] [Google Scholar]
- 21.Gallaher, W. R., J. M. Ball, R. F. Garry, M. C. Griffin, and R. C. Montelaro. 1989. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res. Hum. Retrovir. 5:431-440. [DOI] [PubMed] [Google Scholar]
- 22.Gallaher, W. R., C. DiSimone, and M. J. Buchmeier. 2001. The viral transmembrane superfamily: possible divergence of Arenavirus and Filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh, J. K., and Y. Shai. 1999. Direct evidence that the N-terminal heptad repeat of Sendai virus fusion protein participates in membrane fusion. J. Mol. Biol. 292:531-546. [DOI] [PubMed] [Google Scholar]
- 24.Golding, H., M. Zaitseva, E. de Rosny, L. R. King, J. Manischewitz, I. Sidorov, M. K. Gorny, S. Zolla-Pazner, D. S. Dimitrov, and C. D. Weiss. 2002. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J. Virol. 76:6780-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez, L., Jr., R. A. Brown, D. Richardson, and T. Alber. 1996. Crystal structures of a single coiled-coil peptide in two oligomeric states reveal the basis for structural polymorphism. Nat. Struct. Biol. 3:1002-1009. [DOI] [PubMed] [Google Scholar]
- 26.Harbury, P. B., P. S. Kim, and T. Alber. 1994. Crystal structure of an isoleucine-zipper trimer. Nature 371:80-83. [DOI] [PubMed] [Google Scholar]
- 27.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401-1407. [DOI] [PubMed] [Google Scholar]
- 28.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157:187-253. [DOI] [PubMed] [Google Scholar]
- 29.Jensen, E. M., I. Zelljadt, H. C. Chopra, and M. M. Mason. 1970. Isolation and propagation of a virus from a spontaneous mammary carcinoma of a rhesus monkey. Cancer Res. 30:2388-2393. [PubMed] [Google Scholar]
- 30.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248:20-34. [DOI] [PubMed] [Google Scholar]
- 32.Kouzarides, T., and E. Ziff. 1988. The role of the leucine zipper in the fos-jun interaction. Nature 336:646-651. [DOI] [PubMed] [Google Scholar]
- 33.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 34.Luo, Z., A. M. Matthews, and S. R. Weiss. 1999. Amino acid substitutions within the leucine zipper domain of the murine coronavirus spike protein cause defects in oligomerization and the ability to induce cell-to-cell fusion. J. Virol. 73:8152-8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Shea, E. K., J. D. Klemm, P. S. Kim, and T. Alber. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254:539-544. [DOI] [PubMed] [Google Scholar]
- 36.Ransone, L. J., J. Visvader, P. Sassone-Corsi, and I. M. Verma. 1989. Fos-Jun interaction: mutational analysis of the leucine zipper domain of both proteins. Genes Dev. 3:770-781. [DOI] [PubMed] [Google Scholar]
- 37.Reitter, J. N., T. Sergel, and T. G. Morrison. 1995. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J. Virol. 69:5995-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhee, S. S., H. X. Hui, and E. Hunter. 1990. Preassembled capsids of type D retroviruses contain a signal sufficient for targeting specifically to the plasma membrane. J. Virol. 64:3844-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee, S. S., and E. Hunter. 1987. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J. Virol. 61:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakai, K., S. Dewhurst, X. Y. Ma, and D. J. Volsky. 1988. Differences in cytopathogenicity and host cell range among infectious molecular clones of human immunodeficiency virus type 1 simultaneously isolated from an individual. J. Virol. 62:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skehel, J. J., T. Bizebard, P. A. Bullough, F. M. Hughson, M. Knossow, D. A. Steinhauer, S. A. Wharton, and D. C. Wiley. 1995. Membrane fusion by influenza hemagglutinin. Cold Spring Harbor Symp. Quant. Biol 60:573-580. [DOI] [PubMed] [Google Scholar]
- 43.Stone-Hulslander, J., and T. G. Morrison. 1999. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 73:3630-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White, J. M. 1995. Membrane fusion: the influenza paradigm. Cold Spring Harbor Symp. Quant. Biol. 60:581-588. [DOI] [PubMed] [Google Scholar]
- 46.Wild, C., J. W. Dubay, T. Greenwell, T. Baird, Jr., T. G. Oas, C. McDanal, E. Hunter, and T. Matthews. 1994. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA 91:12676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365-394. [DOI] [PubMed] [Google Scholar]