Abstract
The adenovirus E1B-55K and E4orf6 proteins cooperate during virus infection while performing several tasks that contribute to a productive infection, including the selective nucleocytoplasmic transport of late viral mRNA. Previous studies have shown that the E4orf6 protein retains the E1B-55K protein in the nucleus of human and monkey cells, but not in those of rodents, suggesting that primate-specific cellular factors contribute to the E4orf6-mediated retention of the E1B-55K protein in the nucleus. In an effort to identify these proposed primate-specific cellular factors, the interaction of the E1B-55K and E4orf6 proteins was studied in a panel of stable human-rodent monochromosomal somatic cell hybrids. Analysis of this panel of cell lines has demonstrated the existence of an activity associated with human chromosome 21 that permits the E1B-55K and E4orf6 proteins to colocalize in the nucleus of a rodent cell. Additional hybrid cells bearing portions of human chromosome 21 were used to map this activity to a 10-megabase-pair segment of the chromosome, extending from 21q22.12 to a region near the q terminus. Strikingly, this region also facilitates the expression of adenovirus late genes in a rodent cell background while having little impact on the expression of early viral genes.
The late phase of a productive infection with adenovirus (Ad) type 2 or type 5 is marked by the selective accumulation and efficient translation of late viral mRNAs in the cytoplasm (15, 32). By contrast, most newly synthesized cellular mRNAs are retained in the nucleus during the late stages of infection (2, 41). The ability of Ad to reprogram mRNA transport is mediated by a complex that includes the Ad early region 1B 55-kDa (E1B-55K) and early region 4 open reading frame 6 (E4orf6) proteins (2, 7, 8, 23, 32, 41). These proteins have been shown to exist as a physical complex both in vitro and in vivo (21, 51). In cells infected with mutant viruses that fail to express the E1B-55K or the E4orf6 genes, the export of viral mRNA is impeded while cellular mRNA continues to be transported at near-normal rates (32, 41). Since the phenotype of a double-mutant virus unable to express both the E1B-55K and E4orf6 genes is no more defective than that of either of the single-mutant viruses (8, 12), it was concluded that the E1B-55K-E4orf6 complex was the functional moiety responsible for altering mRNA transport during Ad infection.
The E4orf6 protein localizes to the nucleus both in Ad-infected cells (12) as well as in transiently transfected cells, in which it is evenly distributed throughout the nucleoplasm and is excluded from the nucleoli (6, 21, 38, 39, 57). By comparison, the E1B-55K protein shows a complex distribution within the cytoplasm and nucleus of cells infected with wild-type Ad5 (40, 53). At late times of infection, a portion of the E1B-55K protein is uniformly distributed in the cytoplasm and is also found in a perinuclear body (i.e., adjacent to the nucleus) (33, 60). The majority of the E1B-55K protein, however, is located in the nucleus (40, 52, 53). Within the nucleus, the E1B-55K protein is found in nuclear spicules or tracts and also localizes within and about the periphery of the viral centers (19, 29, 40), which are sites of viral DNA replication and transcription (42). Significantly, at times when late viral mRNA export is maximal, the E4orf6 protein directs the E1B-55K protein to these sites of viral RNA processing (40). By contrast, in cells infected with a mutant virus that fails to express the E4orf6 gene, the E1B-55K protein was found mainly as nuclear spicules and failed to associate with the viral centers (32, 40). Cells infected with a mutant virus unable to express the E4orf6 gene also fail to export late viral messages to the cytoplasm (8, 23, 27, 49, 50). Thus, the proper interaction of the E1B-55K and E4orf6 proteins at the viral centers appears to be a crucial step in the export of late viral mRNAs (15, 19, 48). To account for the alterations in mRNA transport that occur during Ad infection, it was hypothesized that the E1B-55K-E4orf6 complex recruits a limiting cellular factor to the viral centers, leading to the selective export of late viral transcripts to the cytoplasm and the retention of most cellular transcripts in the nucleus (40).
In contrast to its nuclear localization within infected cells, the E1B-55K protein is found in the cytoplasm and not in the nucleus of transiently transfected cells (6, 14, 21, 38, 39). Although it recently has been shown that the E1B-55K protein shuttles between the cytoplasm and the nucleus (30), the protein predominantly resides in the cytoplasm when expressed alone. Studies on the interaction of the E1B-55K and E4orf6 proteins when coexpressed by transient transfection have shown that the E4orf6 protein retains the E1B-55K protein in the nuclei of human and primate cells but not in those of most rodent cells (21). Furthermore, fusion of HeLa cells to rat cells enabled the E4orf6 protein to retain the E1B-55K protein in the rat cell nucleus. Thus, it was suggested that a primate-specific cellular factor or activity mediated the interaction between the E1B-55K and E4orf6 proteins (21). Gabler and associates identified a cellular protein termed E1B-AP5 that binds to the E1B-55K protein in vitro and in Ad-infected cells (18). Interestingly, E1B-AP5 is a nuclear RNA-binding protein that prevents the block on the export of host-cell mRNA and also promotes the increased cytoplasmic accumulation of late viral transcripts. Though it appears that the E1B-AP5 protein functions with the E1B-55K-E4orf6 complex to alter mRNA transport in infected cells, it remains to be shown whether the E1B-AP5 protein can mediate the interaction of the E1B-55K and E4orf6 proteins.
In addition to its role in altering mRNA transport within Ad-infected cells, the E1B-55K-E4orf6 complex also promotes the proteasome-mediated degradation of p53 (9, 36, 37, 43, 44, 54) as well as of the Mre11 complex, which is involved in DNA double-strand break repair (55). Although it remains to be shown whether these multiple functions of the E1B-55K-E4orf6 complex are linked, it is clear that these Ad proteins cooperate with multiple cellular factors within the host cell.
Here, we examine the nuclear colocalization of the E1B-55K and E4orf6 proteins in a panel of highly stable human-rodent monochromosomal somatic cell hybrids (11). We show that an activity associated with human chromosome 21 allows the E4orf6 protein to retain the E1B-55K proteins in the nucleus in a rodent cell background. Regional and sequence-tagged site PCR (STS-PCR) mapping studies have mapped this activity to a 10-Mb region of chromosome 21, extending from region 21q22.12 to region 21q22.3 near the q terminus. Strikingly, this region is necessary both for the nuclear colocalization of the E1B-55K and E4orf6 proteins and for the efficient expression of Ad late viral genes in a rodent cell background.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
Cell culture media, cell culture supplements, and serum were obtained from InVitrogen Life Technologies/GIBCO (Gaithersburg, Md.) through the Tissue Culture Core Laboratory of the Comprehensive Cancer Center of Wake Forest University. HeLa (CCL2.2) cells were maintained in Dulbecco's modified Eagle's minimal essential medium (DMEM) supplemented with 10% newborn calf serum. Mouse A9 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). The A9 monochromosomal somatic cell hybrids (11) were maintained in DMEM supplemented with 10% FBS and 400 U of hygromycin B per ml. Both the GM 10322 and GM 10323 cell lines were maintained in Alpha modification MEM (Eagle) with ribonucleosides and supplemented with 10% FBS. The GM 08854 cell line was maintained in DMEM (high glucose) supplemented with 10% FBS. The GM 14064 cell line was maintained in Ham's F12 medium supplemented with 10% FBS. The GM 14065 cell line was maintained in Ham's F12-deficient (glycine-free) medium supplemented with 50 mg of proline per liter and 5% FBS. Both the GM 11980 and GM 11979 cell lines were maintained in Ham's F12 medium supplemented with 7% FBS. The GM 11752 cell line was maintained in Ham's F12 medium (uridine deficient) supplemented with 50 mg of proline per liter and 5% dialyzed FBS. The GM 14227 cell line was maintained in high-glucose DMEM supplemented with 34.5 mg of proline per liter and 10% FBS. The GM 14416 cell line was maintained in Ham's F12-deficient medium (purine free) supplemented with 50 mg of proline per liter, 3 × 10−5M uridine, and 5% dialyzed FBS. The GM 10157 cell line was maintained in Ham's F12-deficient medium (minus hypoxanthine) supplemented with 0.2 mM proline and 5% dialyzed FBS. The GM 09142 cell line was maintained in Alpha modification MEM (Eagle) with ribonucleosides supplemented with 15% FBS. The GM 14415 cell line was maintained in Ham's F12-deficient medium (glycine free) supplemented with 50 mg of proline per liter and 5% dialyzed FBS. The GM 11130 cell line was maintained in DMEM supplemented with 0.1 mM sodium hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine (InVitrogen Life Technologies) and 10% FBS. The GM 10063 cell line was maintained in DMEM supplemented with 0.1 mM sodium hypoxanthine, 0.4 μM aminopterin, 16 μM thymidine (InVitrogen Life Technologies), and 15% FBS. Chinese hamster ovary (CHO-K1) cells were maintained in 50:50 DMEM-Ham's F12 medium supplemented with 10% FBS. CHO cells stably expressing the human Coxsackie-adenovirus receptor (CAR) gene (i.e., CHO-hCAR) were maintained in Alpha-minus MEM without ribonucleosides and supplemented with 10% dialyzed FBS.
The plasmids bearing the E1B-55K and E4orf6 genes were described previously (21). Herring sperm DNA was used as carrier DNA in transfection experiments. The plasmid encoding the hCAR was generously provided by J. Bergelson of the Division of Immunologic and Infectious Diseases at the Children's Hospital of Philadelphia, and the plasmid was described in reference 3.
The recombinant vaccinia virus used to direct synthesis of the T7 RNA polymerase, vTF7.3, was created by Fuerst et al. (17). Expression of the E1B-55K and E4orf6 genes from the T7 promoter was achieved as described previously (21). Briefly, 105 cells were infected with vTF7.3 in reduced serum medium and transfected with 2 μg of plasmid DNA mixed with 6 μg of Lipofectin (InVitrogen Life Technologies) in accordance with the manufacturer's recommendation. The cells were analyzed by immunofluorescence between 14 and 16 h after infection and transfection.
Ad5 strain dl309 served as the wild-type Ad5 used in these studies. dl309 lacks a portion of the E3 region, which has been shown to be dispensable for growth in culture (28). The virus was propagated in 293 cells (22), and virus stock was prepared by sequential centrifugation through discontinuous and isopycnic CsCl gradients in a Beckman model L70 ultracentrifuge as described previously (21).
Indirect immunofluorescence.
Indirect immunofluorescence was conducted as described previously (40), with the modification that cells were fixed in 2% formaldehyde freshly prepared from paraformaldehyde. Double-label immunofluorescence was performed with the E4orf6 protein-specific mouse monoclonal antibody MAb3 (34) as undiluted hybridoma culture supernatant fluid and the E1B-55K protein-specific rat monoclonal antibody 9C10 (60) at 1 μg per ml (Oncogene Science, Uniondale, N.Y.). The secondary antibodies, used at 2 μg per ml each, were Alexa Flour 488-conjugated goat anti-mouse immunoglobulin G (IgG) and Alexa Fluor 568-conjugated goat anti-rat IgG (Molecular Probes, Eugene, Ore.).
Some samples were analyzed by a Zeiss LSM 510 confocal laser scanning device fitted to an Axioplan 2 microscope with a ×60/NA 1.2 water immersion objective. Alexa Fluor 488 and Alexa Fluor 568 dyes were excited by krypton ion and helium-neon laser excitation, respectively. Single optical sections of approximately 1.5-μm depth at the level of the cytoplasm were recorded as TIFF files by using the LSM 510 software. Other samples were analyzed by epifluorescence microscopy by using a Nikon TE300 inverted microscope fitted with filters appropriate for 4′,6-diamidino-2-phenylindole (DAPI), Alexa Fluor 488, and Alexa Fluor 568 excitation. Images were acquired with a monochrome Retiga EX 1350 digital camera (QImaging Corp., Burnaby, British Columbia, Canada) with a ×63/1.2 NA (dry), ×60/1.4 NA, or ×100/1.4 NA oil immersion objective. The relative brightness and contrast of the digital images within each figure were adjusted to the same extent, and the appropriate colors were assigned with the use of Photoshop 5.5 (Adobe Systems Inc., San Jose, Calif.).
Regional mapping of chromosome 21-associated activity.
To localize the activity encoded on human chromosome 21 that permitted the nuclear colocalization of the E1B-55K and E4orf6 proteins, we obtained a chromosome 21 regional mapping panel from the Coriell Cell Repositories of the National Institute of General Medical Sciences, Camden, N.J. Each of the cell lines in the panel was transfected to transiently express the E1B-55K gene alone or both the E1B-55K and E4orf6 genes by using the recombinant vaccinia virus T7 infection-transfection system described above. The localization of the proteins was then determined by double-label immunofluorescence as described above. Those cell lines in which the frequency of E1B-55K-E4orf6 nuclear colocalization was greater than that measured for the parental cell line were scored as positive.
STS-PCR mapping.
In order to localize the position of the breakpoint within the hybrid cell line GM 14064 carrying the t(8;21) (q22.1;q22.3) translocation, markers along the long arm of chromosome 21 were chosen at regular intervals from the NCBI database (http://www.ncbi.nlm.nih.gov). Genomic DNA was purified from GM 14064 cells and from human and mouse 3T3 cells to provide a positive and negative control. Fifty nanograms of DNA was amplified by PCR with 10 μM of each STS primer pair and Taq polymerase (InVitrogen Life Technologies) by using the recommended buffer conditions. Thirty cycles of amplification were carried out in 50-μl reaction volumes following the sequence of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. The presence and size of the predicted PCR products were determined by electrophoresis through a 2% agarose gel followed by ethidium bromide staining.
Quantification of cells showing E1B-55K-E4orf6 nuclear colocalization.
For quantitative immunofluorescence, cells were transiently transfected by using the recombinant vaccinia virus T7 infection-transfection system to coexpress the E1B-55K and E4orf6 genes. The localization of the proteins was determined by double-label immunofluorescence, and the percentage of cells showing nuclear colocalization of the proteins was determined. Cells were scored as positive for E1B-55K-E4orf6 nuclear colocalization for cells in which at least a portion of the E1B-55K protein was coincident in the nucleus with the E4orf6 protein and in which nucleoli were clearly excluded. The two-tailed Student's t test was used to evaluate differences between paired cell lines.
Heterokaryon formation by PEG fusion.
Mouse A9 cells (4 × 105) grown on 35-mm-diameter culture dishes were infected with the recombinant vaccinia virus, vTF7.3, to express the T7 RNA polymerase gene and transfected with plasmids to express the E1B-55K gene alone or to coexpress the E1B-55K and E4orf6 genes as described above. HeLa cells were harvested by treatment with trypsin and suspended in DMEM with 10% FBS. At 7 h postinfection, the virus-DNA mixture on the A9 cells was removed and replaced with 8 × 105 HeLa cells in 2 ml of solution. The cells were allowed to attach for 1 h at 37°C. Heterokaryons were formed with polyethylene glycol (PEG) (Roche Diagnostics, Pleasanton, Calif.) by the method of Davidson and Gerald (13). Briefly, a 50% solution of approximately 1,500-molecular-weight PEG in 75 mM HEPES (pH 8.0) was warmed to 37°C. At 8 h postinfection, the medium was thoroughly aspirated from the cocultures of A9 and HeLa cells, and 1 ml of the PEG solution was added. After 1 min, the PEG solution was removed and the cells were washed three times with serum-free DMEM, replacing the final wash with DMEM containing 10% FBS and 100 μg of cycloheximide (Sigma Chemical Co., St. Louis, Mo.) per ml. The heterokaryons were returned to 37°C for an additional 4 h before processing for immunofluorescence. Cells were processed for double-label immunofluorescence by using a rat monoclonal antibody, 9C10, specific for the E1B-55K protein (60), and the antibody, N9C1, specific for the 84-kDa subunit of the human Ku protein (BABCO, Richmond, Calif.; 56). The secondary antibodies used were the same as those described above.
Viral gene expression and viral DNA synthesis measurements.
Cells passaged 18 h previously at 2 × 105 per 35-mm-diameter culture dish in a 2-ml volume were transfected with 1 μg of cDNA encoding hCAR by using Lipofectamine Plus (InVitrogen Life Technologies) in accordance with the manufacturer's recommendations. Twenty-four hours after transfection, cells were infected with an effective multiplicity of 1 PFU per cell of wild-type Ad5, dl309. Forty-eight hours after infection, replicate samples were prepared for immunofluorescent evaluation of hCAR, early and late viral gene expression, immunoblot analysis of viral proteins, and slot blot analysis of viral DNA. Antibodies used for immunofluorescence included B6-8 for the E2A DNA-binding protein (DBP) (47); 2Pb-1 for the late protein, penton base (10); and RmcB, specific for hCAR (26). The secondary antibody was Alexa Fluor 488-conjugated goat anti-mouse IgG used as previously described. The stained cells were visualized with a Nikon Eclipse TE300 inverted microscope, and the percentage of cells staining for each Ad protein was determined. CAR staining was used as a measure of transfection efficiency in the rodent cell lines. Staining for DBP provided a measure of the fraction of productively infected cells that typically correlated with the efficiency of CAR transfection in the rodent cell lines. The efficiency of late gene expression was expressed as the ratio of penton base-positive cells to DBP-positive cells.
For immunoblot analysis, the adherent and detached cells were harvested, washed with ice-cold phosphate-buffered saline, and lysed by the addition of freshly prepared sodium dodecyl sulfate (SDS) protein sample buffer (60 mM Tris-Cl, [pH 6.8], 1% SDS, 5 mM EDTA, 50 mM dithiothreitol, and 10% [vol/vol] glycerol) at 2 × 107 cells per ml. The cells were disrupted by sonication, and the lysates were clarified by centrifugation at 14,000 × g for 5 min before separating the proteins from 105 infected cells per lane by SDS-12.5% polyacrylamide gel electrophoresis. The separated proteins were transferred to a nitrocellulose support and visualized by blotting with the E1A-specific M73 antibody and chemiluminescent detection by using the FemtoWest kit as per the manufacturer's recommendations (Pierce, Rockford, Ill.).
Viral DNA present in the infected cell culture was measured by slot blot analysis essentially as described previously (21). Briefly, DNA was isolated from the infected cells by detergent lysis and proteinase K treatment followed by extraction with phenol and chloroform. Serial dilutions of infected cell DNA were immobilized on a nylon membrane (Nytran, Schleicher and Schuell, Keene, N.H.) and then hybridized to radioactively labeled Ad DNA fragments. The amount of bound DNA was quantified by phosphorescence imaging (ImageQuant, Molecular Dynamics, Sunnyvale, Calif.). The amount of Ad DNA per cell was expressed relative to the amount measured in infected HeLa cells.
RESULTS
The E1B-55K and E4orf6 proteins colocalize in the nuclei of HeLa cells but not in the nuclei of mouse A9 cells.
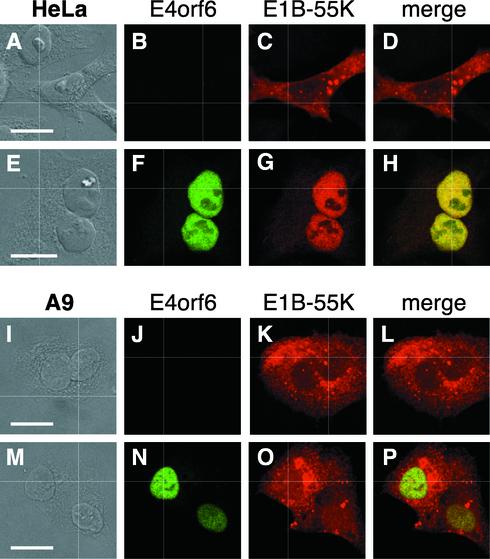
The ability of the E4orf6 protein to direct nuclear localization of the E1B-55K protein in HeLa cells is seen in Fig. 1. HeLa cells were transiently transfected by using the recombinant vaccinia virus T7 infection-transfection system (17) to express the E1B-55K gene alone (Fig. 1A through D) or to coexpress both the E1B-55K and E4orf6 genes (Fig. 1E through H). After transfection, the localization of the E1B-55K and E4orf6 proteins was determined by double-label immunofluorescence with a rat monoclonal antibody that was specific for the E1B-55K protein, 9C10 (60), and a mouse monoclonal antibody that was specific for the E4orf6 protein, MAb3 (34).
FIG. 1.
The E1B-55K and E4orf6 proteins colocalize in the nuclei of HeLa cells but not in mouse cells. HeLa cells (A through H) or mouse A9 cells (I through P) were infected with the recombinant vaccinia virus vTF7.3 to establish expression of the T7 RNA polymerase and were then transfected with cDNA under the control of the T7 promoter to express the E1B-55K gene (A through D, I through L) or to express both the E1B-55K and E4orf6 genes (E through H, M through P). Simultaneous double-label immunofluorescence was performed at 15 h after transfection, and representative cells are shown. (A, E, I, and M) DIC image; (B, F, J, and N) staining for the E4orf6 protein with mouse monoclonal antibody MAb3; (C, G, K, and O) staining for the E1B-55K protein with rat monoclonal antibody 9C10; (D, H, L, and P) merged red and green image. The localization of the proteins was recorded by using confocal laser scanning microscopy. Bars, 10 μm.
Consistent with previous findings (21, 38), the E1B-55K protein was restricted to the cytoplasm and was largely excluded from the nucleus of HeLa cells when expressed in the absence of the E4orf6 protein (Fig. 1A through D). Also as reported previously, the E4orf6 protein was diffusely distributed in the nucleus and was excluded from the nucleoli. When coexpressed with the E1B-55K protein, the E4orf6 protein retained the E1B-55K protein in the HeLa cell nucleus (Fig. 1E through H). By contrast, when the E1B-55K protein (Fig. 1I through L) or both E1B-55K and E4orf6 proteins (Fig. 1 M to P) were synthesized in mouse A9 cells, the E1B-55K protein was found predominantly in the cytoplasm. Together, these results are consistent with the hypothesis that a primate-specific cellular factor mediates the interaction of the E1B-55K and E4orf6 proteins, since the proteins colocalize in human cell nuclei but fail to do so in mouse cell nuclei.
Fusion of human cells to mouse A9 cells restores the ability of the E4orf6 protein to direct nuclear localization of the E1B-55K protein.
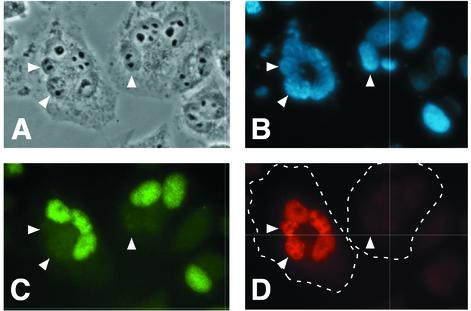
The nuclear colocalization of the E1B-55K and E4orf6 proteins in HeLa cells but not in A9 cells could either reflect a permissive environment in HeLa cells or an inhibitory environment in A9 cells for E1B-55K-E4orf6 interaction. To address this issue, HeLa cells were fused to A9 cells expressing the E1B-55K and E4orf6 genes (Fig. 2). The E1B-55K and E4orf6 proteins were coexpressed in mouse A9 cells by using the recombinant vaccinia virus T7 infection-transfection system. After 8 h, noninfected, nontransfected HeLa cells were fused to the transfected A9 cells by using PEG. The resulting heterokaryons were then allowed to stabilize for 4 h in the presence of cycloheximide to inhibit new protein synthesis. At this time, the localization of the E1B-55K protein and the 84-kDa subunit of the human-specific Ku protein in the heterokaryons was determined by double-label immunofluorescence microscopy.
FIG. 2.
Fusion of human cells to mouse A9 cells expressing the E1B-55K and E4orf6 genes permits the E1B-55K protein to enter mouse A9 cell nuclei. Expression of the E1B-55K and E4orf6 proteins was established in mouse A9 cells with the recombinant vaccinia virus T7 infection-transfection system. HeLa cells were harvested by trypsin treatment and allowed to attach to the substrate with adherent A9 cells. The HeLa and A9 cells were fused 8 h after transfection with PEG. The resulting heterokaryons were maintained in the presence of cycloheximide (100 μg/ml) for 4 h before processing for double-label immunofluorescence. (A) Phase contrast image showing two heterokaryons; (B) DAPI-stained image showing DNA; (C) staining for the large subunit of the human Ku protein specifically labels the HeLa cell nuclei; (D) staining for the E1B-55K protein with the rat monoclonal antibody 9C10. A9 cell nuclei are marked by arrowheads.
The HeLa cell nuclei in the heterokaryons shown in Fig. 2A can be distinguished by their larger size and their larger, more phase-dense nucleoli. By contrast, the A9 cell nuclei within the heterokaryon (indicated by arrowheads) are smaller and also contain smaller nucleoli. The cell nuclei are also readily visualized by the DAPI staining shown in Fig. 2B. The cytoplasmic replication of the recombinant vaccinia virus used to infect the A9 cells is evident by the DAPI-stained, extranuclear DNA. A mouse antibody specific for the large subunit of the human Ku protein, N9C1 (56), was used to identify the HeLa cell nuclei contained within the heterokaryons in Fig. 2C. In Fig. 2D, the E1B-55K protein was found in both the HeLa and A9 cell nuclei contained within the heterokaryon on the left. There was no corresponding E1B-55K staining in the heterokaryon on the right because none of the A9 cells of the neighboring heterokaryon were successfully transfected. The ability of the E1B-55K protein to enter both the HeLa and A9 cell nuclei was dependent on the E4orf6 protein, since the E1B-55K protein failed to localize to A9 cell nuclei of heterokaryons expressing the E1B-55K protein alone (data not shown).
The ability of the E1B-55K protein to localize to mouse A9 cell nuclei upon fusion with HeLa cells supports the notion that human cells provide a permissive environment for E1B-55K-E4orf6 interaction, as opposed to mouse cells, the environments of which are inhibitory to this interaction. Furthermore, since the heterokaryons were maintained in the presence of cycloheximide to inhibit new protein synthesis, these results also suggest that HeLa cells contain a preexisting, diffusible factor or activity that can mediate the interaction of the E1B-55K and E4orf6 proteins. It has recently been shown that the E1B-55K protein is an actively shuttling protein in the absence of the E4orf6 protein (16, 30). Thus, it is possible that the interaction of the E1B-55K and E4orf6 proteins in the heterokaryons results from a HeLa cell-specific factor that acts to retain the E1B-55K-E4orf6 complex in the A9 cell nucleus. Because the E1B-55K and E4orf6 proteins appeared in all nuclei of the heterokaryon, it is also possible that the E1B-55K-E4orf6 protein complex migrated from the mouse cell nucleus to the human cell nucleus in the heterokaryon, as previously suggested (6, 14, 21, 38, 39).
Human chromosome 21 permits the nuclear colocalization of the E1B-55K and E4orf6 proteins in a mouse cell background.
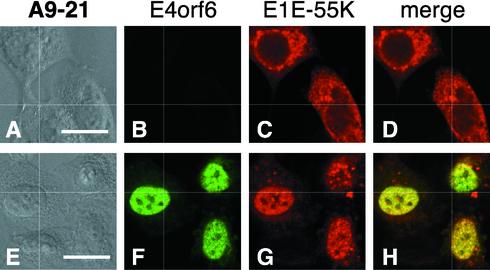
To determine if a single human chromosome could permit nuclear colocalization of the E1B-55K and E4orf6 proteins, we examined the localization of these two proteins in a panel of human-rodent monochromosomal somatic cell hybrids (11). This panel of cell lines was created by using the method of microcell-mediated chromosome transfer in a mouse A9 cell background with normal human diploid fibroblasts as the chromosome donor. The panel contains 23 independent cell lines—one for each of the 22 human autosomes as well as the X chromosome (11). Each of these monochromosomal hybrid cell lines, as well as HeLa cells and A9 cells as positive and negative controls, respectively, was evaluated for E1B-55K-E4orf6 nuclear colocalization. Each cell line was transiently transfected with the recombinant vaccinia virus T7 infection-transfection system described previously to express the E1B-55K gene alone or to coexpress both the E1B-55K and E4orf6 genes. The localization of the proteins was then determined by double-label immunofluorescence. In all 23 hybrid cell lines, the E1B-55K protein was restricted to the cytoplasm and was excluded from the nucleus in the absence of the E4orf6 protein (Fig. 3A through D and data not shown). Strikingly, and in contrast to what was observed in the parental A9 cells (Fig. 1M through P), the E4orf6 protein retained the E1B-55K protein in the nuclei of approximately 50% of the A9-21 hybrid cells (Fig. 3E through H). Therefore, gene products encoded on human chromosome 21 permit E1B-55K-E4orf6 nuclear colocalization in a mouse cell background.
FIG. 3.
Human chromosome 21 permits the nuclear colocalization of the E1B-55K and E4orf6 proteins in a mouse cell background in the A9-21 cell line. The E1B-55K protein was expressed alone (A through D) or coexpressed with the E4orf6 protein (E through H) in A9-21 cells as described in the legend to Fig. 1. (A and E) DIC image; (B and F) staining for the E4orf6 protein with mouse monoclonal antibody MAb3; (C and G) staining for the E1B-55K protein with rat monoclonal antibody 9C10; (D and H) merged red and green image. The localization of the proteins was recorded by using confocal laser scanning microscopy. Bars, 10 μm.
Interestingly, human chromosome 21 encodes the CAR at 21q11.2 (5, 35). CAR is the primary Ad receptor that mediates the attachment of virions to host cells (3, 35) and is the most obvious species-specific Ad-related gene on human chromosome 21. Although it seems unlikely that a cell surface molecule such as CAR could directly mediate the nuclear retention of the E1B-55K protein, it is possible that indirect effects of CAR expression could be involved. Therefore, to determine if CAR could permit E1B-55K-E4orf6 nuclear colocalization, the E1B-55K and E4orf6 genes were transiently expressed in CHO cells that stably expressed the human CAR gene, i.e., CHO-hCAR. CAR did not allow the E4orf6 protein to retain the E1B-55K protein in the nucleus of CHO-hCAR cells (data not shown). These results therefore suggest that chromosome 21 gene products independent of or in addition to CAR are necessary for E1B-55K-E4orf6 interaction.
The E1B-55K-E4orf6-related activity associated with chromosome 21 localizes to a 10-Mb region of the chromosome, extending from 21q22.12 to 21q22.3.
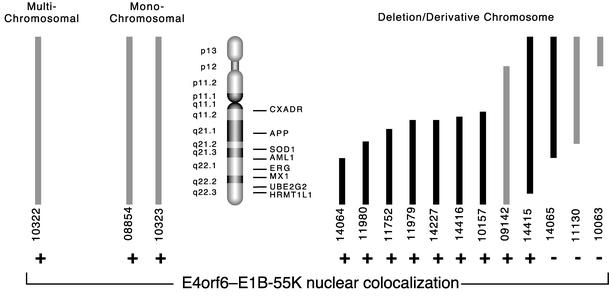
Having determined that an activity associated with human chromosome 21 permits E1B-55K-E4orf6 nuclear colocalization (Fig. 3), we further mapped the activity by screening additional hybrid cell lines that contained various fragments of chromosome 21 for E1B-55K-E4orf6 nuclear colocalization. For these studies, a human chromosome 21 regional mapping panel was obtained from the Coriell Cell Repositories of the National Institute of General Medical Sciences. This panel includes cell lines containing an intact human chromosome 21 as well as cell lines that contained various fragments of the chromosome (Fig. 4). Consistent with the observation that the E1B-55K and E4orf6 proteins colocalize in the nuclei of A9-21 cells, these proteins were also found to colocalize in the nuclei of the hybrid cell lines that contained an intact chromosome 21: GM 10322, GM 08854, and GM 10323 (Fig. 4).
FIG. 4.
An activity encoded in the q-terminal region of chromosome 21 permits E1B-55K-E4orf6 nuclear colocalization. A panel of hamster CHO cells (solid bars) and mouse A9 cells (gray bars) containing fragments of chromosome 21 were obtained from the Coriell Cell Repositories of the National Institute of General Medical Sciences. The E1B-55K and E4orf6 genes were expressed in each of the cell lines by using the recombinant vaccinia virus T7 infection-transfection system, and the localization of both proteins in the cells was determined by double-label immunofluorescence as described in the legend to Fig. 1. At least three independent experiments were scored for nuclear localization of the E1B-55K protein. Cell lines that exhibited significantly greater E1B-55K nuclear localization than the parental A9 or CHO cells are indicated by a “+.” The Coriell repository number for each cell line is indicated below the vertical bar. The vertical bar indicates the approximate portion of chromosome 21 contained in the cell line. The approximate location of several known genes is indicated to the right of the representation of the chromosome. This figure was modified with permission from an image copyrighted by the Coriell Institute for Medical Research, 2001.
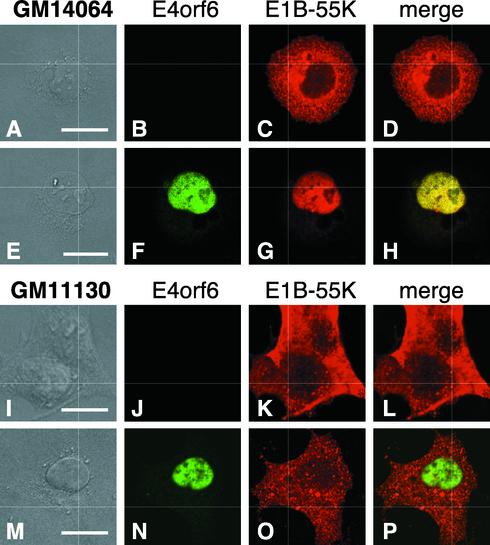
Among the cell lines containing various fragments of chromosome 21, those that contained the q-terminal region of the chromosome showed significant E1B-55K-E4orf6 nuclear colocalization (Fig. 4). The smallest region of chromosome 21 that permitted E1B-55K-E4orf6 interaction was found in the GM 14064 cell line, which contains a q-terminal fragment of chromosome 21 with the proximal breakpoint cytologically mapped to region 21q22.12 (Fig. 4). An example of the extent to which the E4orf6 protein can direct nuclear localization of the E1B-55K protein in the GM 14064 cell line is shown in Fig. 5. Note that the GM 14064 cell line was derived from a CHO cell line, demonstrating that the ability of human chromosome 21 to promote E1B-55K-E4orf6 interaction is not limited to mouse A9 cells.
FIG. 5.
The q terminus of human chromosome 21 permits E1B-55K-E4orf6 nuclear colocalization in a rodent cell background. The E1B-55K gene was expressed alone (A through D, I through L) or coexpressed with the E4orf6 gene (E through H, M through P) in the indicated cell line as described in the legend to Fig. 1. The hamster hybrid cell line GM 14064 contains the q-terminal quarter of human chromosome 21. (A, E, I, and M) DIC image; (B, F, J, and N) staining for the E4orf6 protein with mouse monoclonal antibody MAb3; (C, G, K, and O) staining for the E1B-55K protein with rat monoclonal antibody 9C10; (D, H, L, and P) merged red and green image. The localization of the proteins was recorded by using confocal laser scanning microscopy. Bar, 10 μm.
Cell lines lacking substantial portions of the q-terminal region of chromosome 21 (GM 14065, GM 11130, and GM 10063) were negative for E1B-55K-E4orf6 nuclear colocalization (Fig. 4). A representative example of the inability of the E4orf6 protein to direct nuclear localization of the E1B-55K protein in the GM 11130 cell line is seen in Fig. 5. The failure of the E1B-55K and E4orf6 proteins to colocalize in the nuclei of the GM 11130 cell line (Fig. 5M) also confirmed that the chromosome 21 activity is independent of CAR, which is expressed in the GM 11130 cell line (data not shown).
The smallest fragment of human chromosome 21 that permitted E1B-55K-E4orf6 nuclear colocalization was cytologically mapped to region 21q22.3 through the q terminus. To verify the presence of this chromosomal fragment and to precisely determine the breakpoints of this fragment, STS-PCR mapping was used to define the proximal and distal breakpoints of the chromosome 21 fragment contained in the GM 14064 cell line. The mapping results summarized in Table 1 indicate that the fragment contained in the GM 14064 cell line extends from region 21q22.12 through 21qter. More precisely, the proximal breakpoint of the fragment lies between 32.7 and 32.8 Mb, while the lower limit occurs at the q terminus. Thus, the activity on human chromosome 21 that appears to permit E1B-55K-E4orf6 interaction maps to a region of the chromosome spanning approximately 10 Mb, from 21q22.12 to 21q22.3.
TABLE 1.
STS mapping of the chromosome 21 breakpoint within hybrid GM14064 carrying the t(8;21)(q22.1;q22.3) translocationa
| Marker | Position (Mbp) | Mapb | Resultc |
|---|---|---|---|
| D21S1964 | 24.7 | 21q21.2 | − |
| D21S295 | 25.7 | − | |
| D21S1270 | 28.2 | − | |
| D21S404 | 30.2 | 21q22.1 | − |
| D21S235 | 31.8 | − | |
| D21S1920 | 32.2 | − | |
| D21S1969 | 32.7 | − | |
| STSG48021 | 32.8 | + | |
| D21S1950 | 33.2 | 21q22.12 | + |
| D21S1283 | 33.6 | + | |
| D21S1252 | 34.4 | + | |
| D21S167 | 34.7 | 21q22.13 | ? |
| D21S267 | 35.0 | + | |
| D21S1959 | 36.7 | 21q22.2 | + |
| D21S1259 | 41.8 | 21q22.3 | + |
| D21S2038 | 44.1 | 21q22.3 | + |
Sequence-tagged sites were selected at regular intervals along the long arm of human chromosome 21 (http://www.ncbi.nlm.nih.gov), and their presence in the genomic DNA of GM 14064 was determined by gel electrophoresis of the corresponding PCR products.
If known, the cytogenetic locus associated with the indicated marker is shown.
A PCR reaction product of the predicted size is indicated by “+,” the absence of a product is indicated by “−,” and the failure to obtain a PCR product with any DNA template is indicated by “?.”
An activity associated with the region spanning from 21q22.12 to 21qter increases the number of cells showing E1B-55K-E4orf6 nuclear colocalization.
The fraction of cells showing E1B-55K-E4orf6 nuclear colocalization was quantified in cell lines of varying chromosome 21 status (Fig. 6). For these experiments, the E1B-55K and E4orf6 genes were coexpressed in each of the cell lines by using the recombinant vaccinia virus T7 infection-transfection system and the localization of the proteins was determined by double-label immunofluorescence. Cells containing both E1B-55K and E4orf6 proteins were identified, and the localization of the E1B-55K protein was determined. Cells were scored as positive for nuclear colocalization when E1B-55K-specific staining was coincident with E4orf6-specific staining in the nuclei and in which nucleoli were clearly excluded. A minimum of 300 cells was scored for each cell type in three independent experiments.
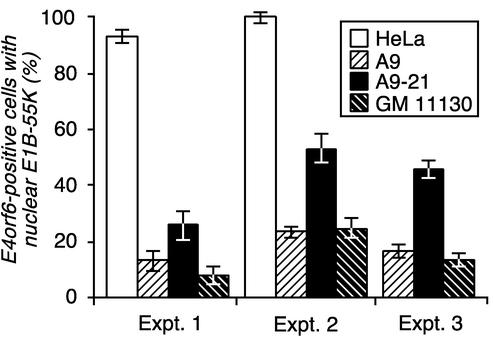
FIG. 6.
An activity contained within the q-terminus of chromosome 21 increases the nuclear colocalization of the E1B-55K and E4orf6 proteins. The E1B-55K and E4orf6 genes were expressed in HeLa cells, mouse A9 cells, or the A9-derived cells, A9-21 and GM 11130 cells, by using the recombinant vaccinia virus T7 infection-transfection system. The cells were then processed for double-label immunofluorescence as described in the legend to Fig. 1. For each independent infection, at least 300 cells containing the E4orf6 protein were scored for nuclear E1B-55K protein and reported as a percentage of the E4orf6-positive cells with the standard deviation (SD) indicated by the error bars. For experiments with one or two independent infections (experiments 1 and 3), the SD was estimated by assuming that the data could be described by a Poisson distribution. The SD associated with experiment 2 was derived from the results of three independent infections.
Although the fraction of cells showing E1B-55K-E4orf6 nuclear colocalization varied between experiments, within each experiment, significantly more A9-21 cells showed nuclear colocalization than the parental A9 cells. On average, the presence of human chromosome 21 in mouse A9-21 cells led to a 2.4-fold increase in the percentage of cells showing E1B-55K-E4orf6 nuclear colocalization—from 19 to 45%. Furthermore, the amount of nuclear E1B-55K protein observed in the 19% of A9 cells showing colocalization was considerably smaller than that observed in the A9-21 cells. In the A9 cells showing colocalization, the majority of the E1B-55K protein remained cytoplasmic, with only a small portion found with the E4orf6 protein in the nucleus. Fewer A9-21 cells, however, showed nuclear colocalization when compared to HeLa cells. An average of 98% of HeLa cells with both the E1B-55K and E4orf6 proteins showed nuclear colocalization. In these cells, nearly complete nuclear retention of the E1B-55K protein was observed. The variable degree to which the E1B-55K protein is retained in the nucleus was previously noted for the interaction between certain E4orf6 variants and the E1B-55K protein (39).
In addition, only 17% of the mouse A9 cell-derived GM 11130 cells showed E1B-55K-E4orf6 nuclear colocalization (Fig. 6). This percentage is indistinguishable from the 19% of parental A9 cells that showed nuclear colocalization (P > 0.7). The GM 11130 cell line contains a fragment of chromosome 21 that lacks the region spanning from 21q21.1 through the q terminus. Thus, chromosome 21 sequences distal to region 21q21.1 appear to contribute to the E1B-55K-E4orf6 interaction observed in a mouse A9 cell background.
The q-terminal portion of human chromosome 21 also promoted E1B-55K-E4orf6 nuclear colocalization in hamster cells. The CHO cell-derived GM 14064 cell line, which contains a region of chromosome 21 extending from 21q22.12 to the q terminus, exhibited E1B-55K-E4orf6 nuclear colocalization in 55% of GM 14064 cells with both Ad proteins (Fig. 5 and data not shown). The CHO cell-derived GM14065 cell line contains the reciprocal chromosome 21 fragment. Unlike the GM 14064 cell line, the frequency of E1B-55K-E4orf6 nuclear colocalization in the GM 14065 cell line was comparable to that seen in the parental CHO cell line (data not shown). Together, results obtained in both mouse and hamster cells suggest that the activity associated with the region from 21q22.12 to 21qter accounts for the entire activity associated with human chromosome 21 that permits E1B-55K-E4orf6 nuclear colocalization.
An activity encoded in region 21q22.12 to 21qter promotes the expression of Ad late viral genes.
Several investigators have suggested that the efficient expression of late Ad genes depends on the proper interaction of the E1B-55K and E4orf6 proteins (19, 21, 40). Whereas human cells are permissive both for E1B-55K-E4orf6 nuclear colocalization and for Ad late gene expression, most rodent cells are negative for nuclear colocalization of the E1B-55K and E4orf6 proteins (21), and most are considered nonpermissive for the expression of Ad late genes (4, 59). Because human chromosome 21 increases the apparent interaction of the E1B-55K and E4orf6 proteins in a mouse A9 cell (Fig. 4) and in a CHO cell background (Fig. 5), we hypothesized that this activity should also promote the efficient expression of late Ad genes in these cells. This was tested by measuring viral DNA replication and the expression of a representative early and late viral gene in Ad-infected cells of varying chromosome 21 status. The results of these experiments, described below and summarized in Table 2, support this hypothesis.
TABLE 2.
Relative frequency of penton base-positive Ad-infected cellsa
| Exptb | Cell line | Chromosome 21 statusc | Penton-positived (%) | ne |
|---|---|---|---|---|
| I | HeLa | ++ | 94 ± 11 | 9, 11 |
| A9 | − | 17 ± 2 | 5, 11 | |
| A9-21 | + | 61 ± 15 | 8, 11 | |
| GM 11130 | 21pter-21q21.1 | 22 ± 3 | 5, 11 | |
| II | HeLa | ++ | 72 ± 24 | 5, 7 |
| GM 14064 | 21q22.12-21qter | 57 ± 11 | 5, 7 | |
| GM 14065 | 21pter-21 q22.12 | 15 ± 1 | 5, 7 |
Cells transiently expressing CAR were infected with a low multiplicity of wild-type Ad. Forty-eight hours after infection, the fraction of cells expressing the E2A DBP and penton base protein was measured by immunofluorescence. The percentage of cells staining for penton base was divided by the percentage of cells staining for the E2A DBP to derive the frequency of infected cells that expressed penton base.
Experiment I compared viral gene expression among HeLa and mouse A9-derived cells. Experiment II compared viral gene expression among HeLa and CHO-derived cells.
The number of copies of human chromosome 21 in the cell line is indicated: ++, two or more copies; +, one copy; −, no human chromosome 21 material. For cell lines that contain a fragment of human chromosome 21, the cytogenetically defined fragment is indicated.
Values are expressed as the percentage ± SD.
The number of the independent infections used to determine the penton base-positive cells and DBP-positive cells, respectively, is indicated.
Some of the rodent cell lines used in these experiments have the gene encoding the Ad receptor, CAR. In addition, the amount of CAR protein varied among each of the cell lines used in this experiment (data not shown). To overcome this complication, all of the cell lines were first transiently transfected to express high levels of the human CAR gene. The efficiency of transfection varied from 10 to 40%. Twenty-four hours after transfection, the cells were infected with an effective multiplicity of 1 PFU (as measured with HeLa or 293 cells) per cell of wild-type Ad, dl309. These conditions, which avoided the use of extraordinarily large amounts of virus, were chosen such that not every CAR-positive cell was expected to be infected. At 48 h postinfection, replicate samples were processed to measure infectivity, viral DNA synthesis, and the synthesis of a representative late protein, the L2 penton base protein; we also evaluated the synthesis of the E1A and E2A DNA-binding proteins as representative early proteins.
Infectivity was determined by immunofluorescent staining for the Ad DBP. Although similar values were obtained by staining for the E1A proteins, the strength of the signal provided by the DBP-specific B6-8 antibody facilitated cell counting. The infectivity levels of the CAR-positive cell lines (HeLa, A9-21, and GM 11130) were similar to and correlated with those of the CAR protein in these cells. The infectivity of the A9 and GM 14064 cell lines, both of which lack the CAR gene, and the infectivity of the GM 14065 cell line, which had low levels of CAR, reflected the efficiency of CAR transfection (data not shown).
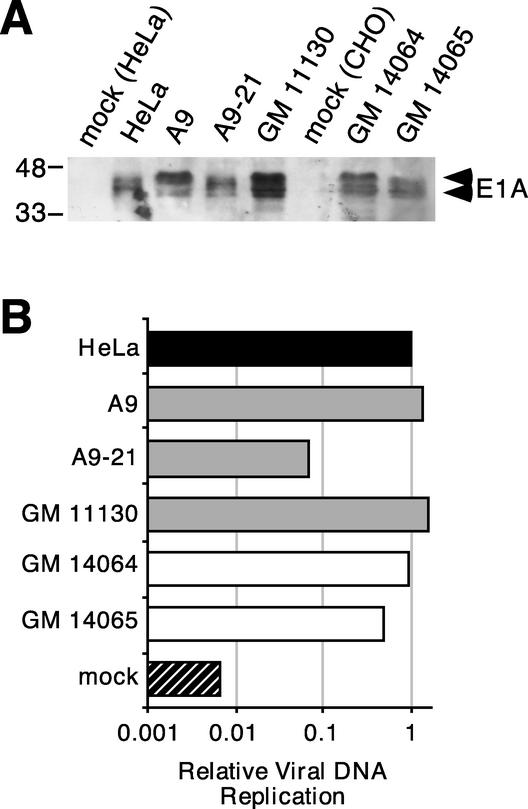
The amount of E1A protein made in the infected cell was apparently not affected by human chromosome 21 sequences (Fig. 7A). Productively infected HeLa cells, A9 cells or A9-21 cells contained comparable amounts of E1A protein at 48 h post infection. The A9-derived GM 11130 cell line contained approximately 1.5- to 2.5-fold more E1A protein per DBP-positive cell than either the A9 cells or HeLa cells did. However, because fewer GM 11130 cells were determined to be DBP positive (approximately 20%), it is possible that more GM 11130 cells were in fact infected but not scored as DBP positive. Most significantly, the infected mouse-derived A9-21 cells contained no more E1A protein than did the infected mouse-derived A9 or GM 11130 cells. Additionally, equivalent amounts of E1A protein were detected in the infected hamster-derived GM 14064 and GM 14065 cells, suggesting that the program of early gene expression in the Ad-infected rodent cells is not significantly influenced by segments of human chromosome 21.
FIG. 7.
Levels of early gene expression and viral DNA synthesis are not affected by chromosome 21 status. Cells transiently expressing high levels of CAR were infected with the wild-type Ad at a multiplicity of approximately 1 and analyzed 48 h after infection for early viral gene expression and viral DNA synthesis. (A) The E1A proteins present in cellular lysates representing equivalent numbers of infected cells were visualized by immunoblotting with the M73 monoclonal antibody (25) and chemiluminescent detection. Only that portion of the membrane containing the two major forms of the E1A proteins is shown. The position of molecular size standards (kDa) is indicated on the left. (B) The amount of Ad DNA present in the cells at 48 h postinfection was determined by hybridizing radioactively labeled Ad DNA to total infected cell DNA immobilized by slot blotting onto a nylon membrane. The amount of Ad-specific hybridization per cell was expressed relative to that measured in the DNA sample from infected HeLa cells. The cell line is indicated on the left of the figure, where “mock” represents the background associated with noninfected HeLa cell DNA. Solid bars, gray bars, and open bars represent HeLa cells, mouse cell lines, and hamster cell lines, respectively. A representative value from two or three independent infections is shown. The values determined from these independent infections typically differed by less than 3% and no more than 12%.
Ad DNA replication in these cell lines was not enhanced by the presence of human chromosome 21 material (Fig. 7B). The amount of viral DNA in HeLa cells 48 h after infection was slightly less than that found in the A9 and GM 11130 mouse cell lines. Somewhat surprisingly, Ad-infected A9-21 cells contained approximately 10-fold less viral DNA than an equivalent number of Ad-infected HeLa cells. Indeed, among all the cell lines studied here, the A9-21 cell line appeared to be the most restrictive toward Ad DNA replication. The amount of newly synthesized viral DNA recovered from the paired hamster-derived cell lines, GM 14064 and GM 14065, was essentially equivalent and comparable to that recovered from HeLa cells. Although approximately 1.5-fold more Ad DNA was recovered from the GM 14064 cell line than from the GM 14065 cell line analyzed in Fig. 7B, this difference could reflect differences in the efficiency of CAR transfection. We noted that although the GM 14065 cell line contains the CAR gene, it was not efficiently expressed in this cell line (data not shown). In summary, human chromosome 21 does not cause significant increases in viral early gene expression or viral genome replication in a rodent cell background, although these experiments do not allow us to rule out a subtle effect on early viral gene expression and viral DNA replication.
Since only a fraction of the cells were productively infected in these experiments, we expected that a comparable or smaller fraction of cells would express late viral genes, represented here by the penton base protein. The relative frequency of penton base-positive cells was derived by normalizing the fraction of cells containing penton base to the fraction of cells with DBP. In the experiments that compared viral gene expression among HeLa and mouse A9-derived cells (Table 2), 94% of the infected HeLa cells contained penton base. By contrast, the penton base protein could be detected in only 17% of the infected A9 cells, which is a 5.5-fold reduction compared to that demonstrated in HeLa cells. In A9-21 cells, approximately 61% of the infected cells expressed the penton base gene; this represents a 3.6-fold increase in expression compared to that in the A9 cells. In the GM 11130 cell line, which lacks the q terminus of chromosome 21, 22% of the infected cells contained penton base protein (Table 2). This value is comparable to the 17% penton base gene expression observed in the parental A9 cell line, suggesting that the q terminus of chromosome 21 is needed for efficient late gene expression in the mouse A9 cell.
To determine if Ad late gene expression correlated with E1B-55K-E4orf6 nuclear colocalization in hamster-derived cell lines, early and late viral gene expression was measured in the GM 14064 and GM 14065 cell lines as described above (Table 2). The E1B-55K and E4orf6 proteins colocalized in the nuclei of 55% of CHO-derived GM 14064 cells, whereas the frequency of colocalization in the nucleus of the GM 14065 cell line was indistinguishable from that of the parental CHO cell line (Fig. 5; data not shown). Correspondingly, 57% of the infected GM 14064 cells synthesized penton base, while only 15% of the infected GM 14065 cells synthesized penton base (Table 2). Thus, approximately fourfold more infected GM 14064 cells showed expression of a representative late Ad gene than infected GM 14065 cells.
Together, these results led us to conclude that an activity encoded on chromosome 21 from the 21q22.12 to 21qter region can promote late Ad gene expression in both a mouse A9 cell and a hamster cell background (Table 2) independently of any effects on early gene expression and viral DNA synthesis. It seems likely that this chromosome 21-associated activity promotes both the nuclear colocalization of the E1B-55K and E4orf6 proteins (Fig. 6) and the efficient expression of late Ad genes, measured here by the synthesis of the penton base protein.
DISCUSSION
In this study, we demonstrated that an activity associated with human chromosome 21 promotes both the nuclear colocalization of the Ad5 E1B-55K and E4orf6 proteins and the expression of late Ad genes in a mouse and a hamster cell background. Furthermore, this chromosome 21-associated activity has been mapped to a 10-Mb region of the q arm of the chromosome, extending from region 21q22.12 to 21q22.3 near the q terminus.
The ability of the E4orf6 protein to direct nuclear localization of the E1B-55K protein was quantified by determining the fraction of cells showing nuclear E1B-55K protein in the presence of the E4orf6 protein. When applied to mouse and hamster cell lines containing various fragments of human chromosome 21, this approach led us to conclude that all of the chromosome 21-associated activity that promotes the apparent E1B-55K-E4orf6 protein interaction is encoded in the q-terminal region of chromosome 21. For example, the E4orf6 protein retained the E1B-55K protein in the nucleus of mouse A9-21 cells, which contain a single intact copy of chromosome 21, to the same extent as in the hamster GM 14064 cells, which contain a portion of the chromosome from 21q22.12 through the q terminus of the chromosome. By contrast, rodent cell lines containing more proximal fragments of human chromosome 21 failed to show increased nuclear colocalization of the E1B-55K and E4orf6 proteins compared to the parental rodent cell lines. Therefore, we conclude that an activity mapping to region 21q22.12 through the q terminus of chromosome 21 is required for the increased nuclear colocalization of E1B-55K and E4orf6 in these rodent cell lines.
The q-terminal region of chromosome 21 carries several genes that are either directly or indirectly related to the E1B-55K and E4orf6 proteins. For example, MX1 at 21q22.3 is an interferon-inducible gene (1) whose mRNA escapes the block on the export of host messages during the late stages of Ad infection (58). The gene UBE2G2, which encodes a ubiquitin-conjugating enzyme, is also found at 21q22.3. The E1B-55K and E4orf6 proteins have recently been shown to exist as part of a Cullin-containing complex that functions as an E3 ubiquitin ligase to target p53 for ubiquitination and, ultimately, for degradation (24, 43). As reported by Querido et al. (43), the identity of the E2 ubiquitin-conjugating enzyme within this complex remains unknown. Perhaps the ubiquitin-conjugating enzyme encoded by UBE2G2 is part of the E1B-55K-E4orf6-Cullin-containing complex described by these two groups. This region of chromosome 21 also includes the gene HRMT1L1, which encodes an hnRNP arginine methyltransferase (31). HRMT1L1 has recently been shown to bind the E1B-55K-associated protein, E1B-AP5, both in vitro and in vivo (31). E1B-AP5 was previously shown by Gabler et al. (18) to bind the E1B-55K protein and to play a role in nucleocytoplasmic mRNA transport in Ad-infected cells. Although the region of chromosome 21 extending from 21q22.12 to the q terminus carries several genes whose products are related to the E1B-55K and E4orf6 proteins, their role in the colocalization of these proteins has yet to be demonstrated. Our unpublished results suggest that the MX1 and HRMT1L1 genes are not sufficient to permit E1B-55K-E4orf6 nuclear colocalization, and additional candidate genes are being tested.
Although an activity associated with the q-terminal portion of human chromosome 21 promotes the colocalization of the E1B-55K and E4orf6 proteins in the rodent cell nucleus, this activity is not sufficient to restore this apparent interaction to the same extent as that observed in human cells. It is possible that the monochromosomal somatic cell hybrids show a lower percentage of E1B-55K-E4orf6 nuclear colocalization because of a gene dosage effect. It is curious that HeLa cells, which contain at least two copies of chromosome 21, showed approximately twofold more (from 45 to 98%) E1B-55K-E4orf6 nuclear colocalization than A9-21 cells, which contain only one copy of chromosome 21. The variation in colocalization observed between the rodent cell lines could also reflect differences in the expression and accumulation of the relevant human gene products. It is also possible that cellular factors encoded by other human chromosomes contribute to the apparent interaction of the E1B-55K and E4orf6 proteins. In support of this possibility, we have observed a low frequency of E1B-55K-E4orf6 nuclear colocalization in four other monochromosomal hybrid mouse cell lines that contained human chromosomes 11, 13, 15, and 16 (data not shown).
The selective export of late Ad mRNA during an infection appears to require the proper interaction of the E1B-55K and E4orf6 proteins in the nucleus of the infected cell. At late stages of an infection, the E4orf6 protein directs the E1B-55K protein to sites of viral transcription and RNA processing (19, 40). Gonzalez and Flint (19) reported that efficient export of penton base mRNA to the cytoplasm correlated with the colocalization of the E1B-55K and E4orf6 proteins at the viral centers in the cell nucleus. One would predict, therefore, that a cellular activity that promotes nuclear colocalization of the E1B-55K and E4orf6 proteins may also facilitate expression of late Ad genes. Indeed, there is a striking correlation between E1B-55K-E4orf6 nuclear colocalization and Ad late gene expression among the various cell lines analyzed in this study. For example, essentially all (>98%) transfected HeLa cells showed E1B-55K-E4orf6 nuclear colocalization, and nearly all (94%) infected HeLa cells contained penton base protein. In the mouse A9-21 cell line, 45% of transfected cells showed E1B-55K-E4orf6 nuclear colocalization, and 61% of infected cells were positive for penton base protein. Moreover, this correlation extends into those rodent cells in which E1B-55K-E4orf6 nuclear colocalization was observed infrequently. In the A9 and GM 11130 cell lines, slightly less than 20% of the transfected cells were positive for nuclear colocalization, while penton base protein was detected in approximately 20% of the infected cells.
The correlation between nuclear colocalization of the E1B-55K and E4orf6 proteins following transfection and Ad late gene expression following infection suggests that these activities may be related. However, the experiments to measure colocalization and the experiments to measure late gene expression were performed separately. Therefore, it is possible that a subset of the transfected cells allowed E1B-55K-E4orf6 nuclear colocalization and that this same subset of cells would have permitted late Ad gene expression if they had been infected. If so, it will be of considerable interest to determine the molecular basis by which a given cell in the population can be predisposed to permitting both E1B-55K-E4orf6 interaction and late gene expression or restricting both activities.
It seems likely that some activities of the E1B-55K-E4orf6 protein complex do not require significant colocalization of these proteins in the cell nucleus. For example, when expressed by transfection in mouse fibroblast cell lines, the bulk of the E1B-55K and E4orf6 proteins remain in separate cellular compartments (21). However, the E1B-55K and E4orf6 proteins direct the degradation of p53 in mouse embryo fibroblasts (45). Efficient late viral gene expression may also occur in the absence of any measurable interaction between the E1B-55K and E4orf6 proteins. For example, E1B-55K-null viruses express late genes at nearly wild-type levels in cells maintained at elevated temperatures (20). We previously described a class of E4orf6 protein variants that appear to support the growth of an E4-mutant virus in an E1B-55K-independent manner (39). We suggested that these E4orf6 protein variants may be able to contribute some of the function normally provided by the E1B-55K-E4orf6 protein complex. Perhaps the requirement for a physical complex of the E1B-55K and E4orf6 proteins can be mitigated by the presence or activity of particular cellular factors.
We previously observed that nuclear colocalization of the E1B-55K and E4orf6 proteins correlated with the permissivity of the cells to Ad replication (21). However, the yield of virus from A9-21 cells was not significantly greater than the yield from comparably infected A9 cells (data not shown), suggesting that formation of the E1B-55K-E4orf6 protein complex and enhanced expression of at least some of the late viral genes is not sufficient to overcome the block to human Ad replication in mouse cells. A similar conclusion was reached in a study by Radna and associates (46) that tried to determine if permissivity to Ad replication was linked to a particular human chromosome in CHO-human hybrid cell lines. Although these investigators found that the presence of human chromosome 3 correlated with restoration of fiber synthesis in these cells, assignment of permissivity to a single human chromosome could not be made. Thus, it appears likely that multiple cellular activities are required for a productive Ad infection. As previously noted, we have detected a low frequency of E1B-55K-E4orf6 nuclear colocalization in the additional monochromosomal somatic cell hybrids containing human chromosomes 11, 13, 15, and 16. Perhaps activities encoded on these chromosomes are also required for the efficient replication of Ad. Interestingly, these chromosomes encode several cellular proteins that have recently been reported to form a complex with the E1B-55K and E4orf6 proteins, including NuMA and Cul5 on chromosome 11, Importin α-1 on chromosome 16, and pp32 on chromosome 15 (24, 43). It will be of interest to determine if these or other activities encoded on these additional chromosomes promote the efficient replication of Ad.
In conclusion, these results suggest that an activity encoded on human chromosome 21 permits the nuclear colocalization of the E1B-55K and E4orf6 proteins and promotes the expression of late Ad genes in a rodent cell background. This activity maps to a 10-Mb region of chromosome 21, extending from region 21q22.12 to region 21q22.3. The identity of the cellular factor encoded in this region and its role in the activity of the E1B-55K-E4orf6 protein complex is currently under investigation.
Acknowledgments
This work was supported in part by Public Health Service grant CA 77342 from the National Cancer Institute and a collaborative research grant (064742/Z/01/Z) from the Wellcome Trust (London, United Kingdom). Tissue culture reagents and services were provided by the Tissue Culture Core Laboratory, and confocal laser scanning microscopy was performed through the Micromed facility, both of which are services of the Comprehensive Cancer Center of Wake Forest University, which is supported in part by the National Cancer Institute grant CA 12197. During part of this work, A.M.C.-M. was supported by a graduate fellowship of the National Science Foundation.
We gratefully acknowledge the expertise and help of Mark Pettenati for FISH analysis. We thank Ken Grant of the Micromed facility for assistance with confocal laser scanning microscopy, Kuie Pao Tsai for technical assistance, and Robin Shepard, Doug Lyles, Steven Mizel, Griff Parks, and Mark Willingham for providing valuable advice on the work in progress and on manuscript preparation. We also thank Paul Freimuth and Phil Gallimore for helpful suggestions at the onset of this work.
REFERENCES
- 1.Aebi, M., J. Fah, N. Hurt, C. E. Samuel, D. Thomis, L. Bazzigher, J. Pavlovic, O. Haller, and P. Staeheli. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 9:5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell, Jr. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 4.Blair, G. E., S. C. Dixon, S. A. Griffiths, and M. E. Zajdel. 1989. Restricted replication of human adenovirus type 5 in mouse cell lines. Virus Res. 14:339-346. [DOI] [PubMed] [Google Scholar]
- 5.Bowles, K. R., J. Gibson, J. Wu, L. G. Shaffer, J. A. Towbin, and N. E. Bowles. 1999. Genomic organization and chromosomal localization of the human Coxsackievirus B-adenovirus receptor gene. Hum. Genet. 105:354-359. [DOI] [PubMed] [Google Scholar]
- 6.Boyer, J. L., and G. Ketner. 2000. Genetic analysis of a potential zinc-binding domain of the adenovirus E4 34k protein. J. Biol. Chem. 275:14969-14978. [DOI] [PubMed] [Google Scholar]
- 7.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 8.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cepko, C. L., P. S. Changelian, and P. A. Sharp. 1981. Immunoprecipitation with two-dimensional pools as a hybridoma screening technique: production and characterization of monoclonal antibodies against adenovirus 2 proteins. Virology 110:385-401. [DOI] [PubMed] [Google Scholar]
- 11.Cuthbert, A. P., D. A. Trott, R. M. Ekong, S. Jezzard, N. L. England, M. Themis, C. M. Todd, and R. F. Newbold. 1995. Construction and characterization of a highly stable human:rodent monochromosomal hybrid panel for genetic complementation and genome mapping studies. Cytogenet. Cell. Genet. 71:68-76. [DOI] [PubMed] [Google Scholar]
- 12.Cutt, J. R., T. Shenk, and P. Hearing. 1987. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J. Virol. 61:543-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson, R. L., and P. S. Gerald. 1976. Improved techniques for the induction of mammalian cell hybridization by polyethylene glycol. Somatic Cell Genet. 2:165-176. [DOI] [PubMed] [Google Scholar]
- 14.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobner, T., and J. Kzhyshkowska. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25-54. [DOI] [PubMed] [Google Scholar]
- 16.Dosch, T., F. Horn, G. Schneider, F. Kratzer, T. Dobner, J. Hauber, and R. H. Stauber. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 75:5677-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA. 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabler, S., H. Schutt, P. Groitl, H. Wolf, T. Shenk, and T. Dobner. 1998. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 72:7960-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, R. A., and S. J. Flint. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodrum, F. D., and D. A. Ornelles. 1998. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J. Virol. 72:9479-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodrum, F. D., T. Shenk, and D. A. Ornelles. 1996. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J. Virol. 70:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 23.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow, E., B. R. Franza, Jr., and C. Schley. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55:533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu, K. H., K. Lonberg-Holm, B. Alstein, and R. L. Crowell. 1988. A monoclonal antibody specific for the cellular receptor for the group B coxsackieviruses. J. Virol. 62:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 29.Konig, C., J. Roth, and M. Dobbelstein. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B-55-kilodalton protein. J. Virol. 73:2253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kratzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19:850-857. [DOI] [PubMed] [Google Scholar]
- 31.Kzhyshkowska, J., H. Schutt, M. Liss, E. Kremmer, R. Stauber, H. Wolf, and T. Dobner. 2001. Heterogeneous nuclear ribonucleoprotein E1B-AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem. J. 358:305-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leppard, K. N., and T. Shenk. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maheswaran, S., C. Englert, S. B. Lee, R. M. Ezzel, J. Settleman, and D. A. Haber. 1998. E1B 55K sequesters WT1 along with p53 within a cytoplasmic body in adenovirus-transformed kidney cells. Oncogene 16:2041-2050. [DOI] [PubMed] [Google Scholar]
- 34.Marton, M. J., S. B. Baim, D. A. Ornelles, and T. Shenk. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J. Virol. 64:2345-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr, G. A., and P. Freimuth. 1997. A single locus on human chromosome 21 directs the expression of a receptor for adenovirus type 2 in mouse A9 cells. J. Virol. 71:412-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 93:11295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 2000. Two distinct activities contribute to the oncogenic potential of the adenovirus type 5 E4orf6 protein. J. Virol. 74:5168-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orlando, J. S., and D. A. Ornelles. 1999. An arginine-faced amphipathic alpha helix is required for adenovirus type 5 E4orf6 protein function. J. Virol. 73:4600-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orlando, J. S., and D. A. Ornelles. 2002. E4orf6 variants with separate abilities to augment adenovirus replication and direct nuclear localization of the E1B 55-kilodalton protein. J. Virol. 76:1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puvion-Dutilleul, F., J. Pedron, and C. Cajean-Feroldi. 1984. Identification of intranuclear structures containing the 72K DNA-binding protein of human adenovirus type 5. Eur. J. Cell Biol. 34:313-322. [PubMed] [Google Scholar]
- 43.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Querido, E., M. R. Morrison, H. Chu-Pham-Dang, S. W. Thirlwell, D. Boivin, P. E. Branton, and M. R. Morisson. 2001. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radna, R. L., B. Foellmer, L. A. Feldman, U. Francke, and H. L. Ozer. 1987. Restriction of human adenovirus replication in Chinese hamster cell lines and their hybrids with human cells. Virus Res. 8:277-299. [DOI] [PubMed] [Google Scholar]
- 47.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480-484. [DOI] [PubMed] [Google Scholar]
- 48.Rubenwolf, S., H. Schutt, M. Nevels, H. Wolf, and T. Dobner. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 71:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandler, A. B., and G. Ketner. 1989. Adenovirus early region 4 is essential for normal stability of late nuclear RNAs. J. Virol. 63:624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandler, A. B., and G. Ketner. 1991. The metabolism of host RNAs in cells infected by an adenovirus E4 mutant. Virology 181:319-326. [DOI] [PubMed] [Google Scholar]
- 51.Sarnow, P., P. Hearing, C. W. Anderson, D. N. Halbert, T. Shenk, and A. J. Levine. 1984. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J. Virol. 49:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510-517. [DOI] [PubMed] [Google Scholar]
- 53.Smiley, J. K., M. A. Young, C. C. Bansbach, and S. J. Flint. 1995. The metabolism of small cellular RNA species during productive subgroup C adenovirus infection. Virology 206:100-107. [DOI] [PubMed] [Google Scholar]
- 54.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16:349-357. [DOI] [PubMed] [Google Scholar]
- 55.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 56.Wang, J., C. H. Chou, J. Blankson, M. Satoh, M. W. Knuth, R. A. Eisenberg, D. S. Pisetsky, and W. H. Reeves. 1993. Murine monoclonal antibodies specific for conserved and non-conserved antigenic determinants of the human and murine Ku autoantigens. Mol. Biol. Rep. 18:15-28. [DOI] [PubMed] [Google Scholar]
- 57.Weigel, S., and M. Dobbelstein. 2000. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J. Virol. 74:764-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, U. C., W. Huang, and S. J. Flint. 1996. mRNA export correlates with activation of transcription in human subgroup C adenovirus-infected cells. J. Virol. 70:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Younghusband, H. B., C. Tyndall, and A. J. Bellett. 1979. Replication and interaction of virus DNA and cellular DNA in mouse cells infected by a human adenovirus. J. Gen. Virol. 45:455-467. [DOI] [PubMed] [Google Scholar]
- 60.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44-58. [DOI] [PubMed] [Google Scholar]