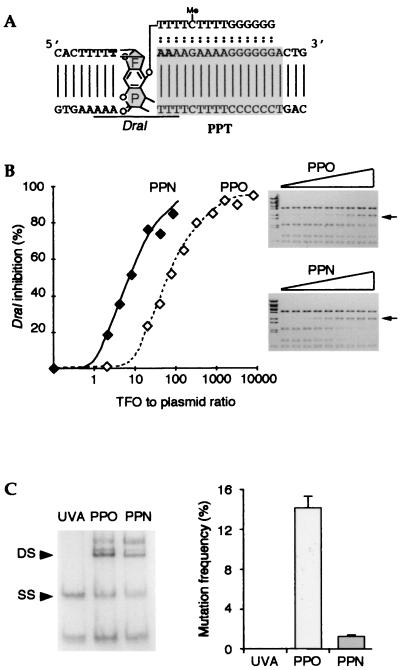
Figure 1.
Analysis of TFOs in vitro and on exogenous plasmids. (A) Sequences of the target gene and the TFO (shown bound on the target gene); two variants of the TFO were used with identical length and sequence but differing in their chemical modification (see text). The ochre codon is indicated in bold. The cytosine in the oligonucleotide is methylated. The furane (F) and pyrone (P) sides of the adduct are indicated, as is the position of the DraI restriction site. (B) The PPN TFO binds to the target sequence with greater affinity than does the PPO TFO. A plasmid including the target sequence (YEplac181_ura3∷hiv1pur) was incubated with increasing doses of TFOs, irradiated with near UV light, and submitted to DraI restriction. Shown are dose curves of DraI inhibition by the PPN or the PPO oligonucleotides, as indicated. (C) Frequency of the mutations generated on an exogenous episomal plasmid (YEplac112_ura3∷hiv1pyr) by the PPO TFO, the PPN TFO, or in the absence of TFO (UVA). Triple helices were preformed on plasmids in vitro. Samples were irradiated, resulting in the introduction of covalent crosslinks into a high proportion of the target plasmids—proportions, which, under the conditions used, were similar for both TFOs as shown by renaturing gel electrophoresis analysis (Left; DS, double-stranded, crosslinked species; SS, single-stranded, noncrosslinked species). The plasmids were used to transform ura3–52 yeast cells. The mutation frequency was defined as the ratio between the number of induced URA3+ revertants and the total number of transformants. Shown is the mean value of three independent experiments.