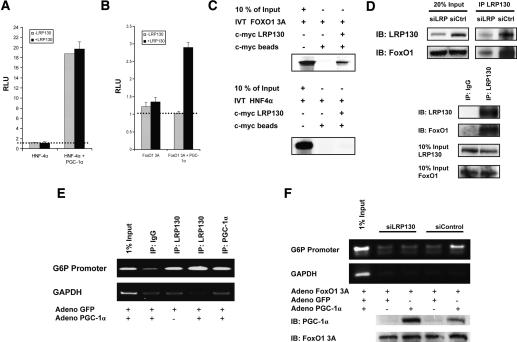
Figure 6.
Effect of LRP130 on coactivation and docking of PGC-1α. H2.35 cells stably depleted of LRP130 were used for the coactivation studies. Exogenous LRP130 was used to restore LRP130 where indicated. (A) LRP130 was cotransfected with PGC-1α and HNF-4α as indicated, and a multimerized AF-1-binding element reporter construct was used. (B) LRP130 was cotransfected with PGC-1α and FoxO1 3A (constitutively active mutant) as indicated. A multimerized IRS-1-binding element reporter construct was used. The dotted line represents reporter construct alone. No statistical difference was observed between reporter alone versus reporter plus transcription factor, or for LRP130 and PGC-1α plus reporter. (C) Assessment of interactions between LRP130 and FoxO1 or HNF4α. FoxO1 or HNF4α was 35S-labeled and incubated with purified full-length S-tagged LRP130 protein. (D) Specific interaction of LRP130 with FoxO1 in liver and cells. (Top panel) Coimmunoprecipitation of LRP130 and FoxO1 from liver whole-cell extract showing dose effect of interaction. (Bottom panel) Coimmunoprecipitation from nuclear extract using H2.35 cells. (E) ChIP showing recruitment of LRP130 to the FoxO1-binding site in PGC-1α knockout (KO) primary hepatocytes. (Top panel) Radioactive ChIP processed by PhosphorImager. (F) ChIP in H2.35 cells stably depleted of LRP130 using an RNAi construct compared with a siControl cell line. The panels below indicate protein levels of PGC-1α and FoxO1 3A. Note, the upper specific band migrates in close proximity to the lower single-peak unreacted primer. Experiments were performed in n ≥ 3 independent biological experiments.