Abstract
Fecal samples from healthy children under 2 years of age living in Berlin, Germany (205 infants), and Melbourne, Australia (184 infants), were investigated for the presence of attaching and effacing (AE) Escherichia coli (AEEC) strains by screening for eae (intimin) genes. Twenty-seven AEEC strains were isolated from 14 children (7.6%) from Melbourne and from 12 children (5.9%) from Berlin. The 27 AEEC strains were classified as enterohemorrhagic E. coli (one strain, producing Shiga toxin 1), typical enteropathogenic E. coli (EPEC) (one strain carrying an EPEC adherence factor [EAF] plasmid), and atypical EPEC (25 strains negative for Shiga toxins and EAF plasmids). The AEEC were divided into 18 different serotypes, O-nontypeable and O-rough strains. Typing of their intimin genes revealed the presence of intimin α in 6 strains, intimin β in 11 strains, intimin γ in 7 strains, intimin ζ in 2 strains, and intimin η in one strain. Analysis of HEp-2 cell adherence showed diffuse adherence or localized adherence-like patterns in 26 AEEC strains; local adherence was found only with the EAF-positive strain. Ten AEEC strains showed an AE property with the fluorescent actin staining (FAS) test. The introduction of an EAF plasmid (pMAR7) converted 11 FAS-negative AEEC strains to FAS positive and increased the FAS reaction in six FAS-positive AEEC strains, indicating that the genes needed for the AE phenotype were functional in these strains. Our finding indicates that atypical EPEC strains could play a double role as strains that naturally immunize against intimin in humans and as reservoirs for new emerging human pathogenic EPEC strains.
The ability to cause attaching and effacing (AE) lesions in cells of the intestinal mucosa was identified as an important pathogenicity factor of enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) strains (42, 64). The AE lesion is generated by intimate attachment of bacteria to the enterocytes, which is followed by aggregation of the cytoskeletal actin and effacement of microvilli (34). Actin aggregation caused by AE E. coli (AEEC) strains results in a typical picture which can be visualized by a fluorescent actin staining (FAS) test which is used for detection of EPEC and EHEC (34, 35, 58). The genes coding for the AE lesion are located on a pathogenicity island termed “locus of enterocyte effacement” (LEE) in EPEC and EHEC strains. The 34-kb core region of the LEE is highly conserved in human and animal EPEC and EHEC strains and contains polycistronic operons harboring genes for type III secretion and adhesion proteins (18, 40, 51, 68). Intimate attachment of bacteria to the eucaryotic cell is mediated by intimin, the product of the eae gene, which is incorporated in the bacterial membrane. Intimin binds to the translocated intimin receptor (Tir), which is presented on the surface of the eucaryotic target cell (for a review, see reference 64). The detection of the intimin gene was taken as an indicator for the presence of functional LEE genes because it was shown that the presence of the eae gene correlated well with a positive FAS assay in most EPEC and EHEC isolates (34, 35, 50).
The characterization of the eae genes of EPEC and EHEC revealed the existence of intimin variants. At present, 10 genetic variants of the eae gene have been identified by nucleotide sequencing and are designated with letters of the Greek alphabet (1, 48, 60, 67). Different variants of the Tir molecule have also been described (15, 33, 49), and the different intimins and tir products are determinants of host tissue tropism and not fully functionally interchangeable between EPEC and EHEC strains (19, 38, 52).
EPEC and EHEC were also shown to be different for their mechanism of adherence to intestinal cells (22, 42). EPEC colonizes the small intestinal mucosa using bundle-forming pili (BFP) as the initial colonization factor. BFP are encoded by EPEC adherence factor (EAF) plasmids which are present in typical EPEC but absent in EHEC strains (42, 43). EAF plasmids are a family of related plasmids which carry genes for BFP (bfp operon) and the perABC (bfpTVW) operon (plasmid-encoded regulator) genes (23, 24, 39, 43, 46, 61). BFP mediate localized adherence (LA) of bacteria to the host cells, and the genes of the per operon are positive regulators of plasmid-encoded bfp genes and chromosomally encoded genes of the LEE, including eae (23, 24, 26, 41, 61). Atypical EPEC strains were described as those that carry the LEE genes but are negative for EAF plasmids, BFP, and Shiga toxin production (63). Atypical EPEC strains do not show LA but show diffuse adherence (DA) or LA-like (LAL) patterns with regard to epithelial cells, which was associated with the absence of BFP and per genes in these strains (50, 54). Similarly, EHEC strains which colonize the large intestine are negative for BFP and do not show LA to cultured mammalian cells. (42).
The investigation of EPEC and EHEC for their intimin types has shown that intimin α is present in strains belonging to classical EPEC serotypes (O55:H6, O127:H6 and O142:H6) whereas intimins γ, ɛ, and θ are present in classical EHEC (O103:H2, O111:NM, O145:NM and O157:[H7]), but not in classical EPEC strains. In contrast, intimin β is found in both classical EPEC (O86:H34, O114:H2, and O128:H2) and EHEC (O26:[H11] and O118:H16) strains (48, 63). Furthermore, intimins α, β, and γ were found to be associated with atypical EPEC strains (63). The association of other intimin types with the different groups of EPEC strains is less known (67).
In this work, we were interested in the frequency and in the virulence characteristics of eae-positive E. coli from children who had no symptoms of gastrointestinal illness. For this, we have investigated 482 fecal E. coli isolates from 389 infants living in the cities of Berlin, Germany, and Melbourne, Australia. The eae-positive E. coli strains from infants were investigated for their relationship to described EPEC and EHEC strains, for their eae gene (intimin) variants by genotyping, and for HEp-2 cell-adherence and the expression of LEE-associated genes by the FAS assay.
MATERIALS AND METHODS
Infants.
Fecal specimens from randomly selected infants less than 2 years old, who had not had any gastrointestinal symptoms for at least 1 week prior to the collection of the specimen and who were currently not taking any antibiotic therapy, were collected in Berlin, Germany, between 1986 and 1987 (205 infants) and Melbourne, Australia, between 1988 and 1994 (184 infants) (5).
Isolation and characterization of E. coli from stool samples.
About 1 g of feces was suspended in 10 ml of nutrient broth (Oxoid, Basingstoke, United Kingdom) and a loopful was spread onto Endo-Agar (Merck, Darmstadt, Germany) (Berlin group) or MacConkey agar (Oxoid) (Melbourne group). After overnight growth at 37°C, 10 single coliform colonies were selected, such that representatives of each type of colonial morphology were examined. The single colony isolates were characterized as E. coli on the basis of their reactions in triple-sugar-iron agar (Oxoid), motility-indole-ornithine medium (Oxoid), o-nitrophenyl-β-d-galactopyranoside (Oxoid), and urease broth (Oxoid). Strains which conformed to being E. coli were stored frozen in 45% glycerol at −70°C.
Serotype identification.
The E. coli strains were O-antigen and H-antigen serotyped using previously described methods (6, 47). Nonmotile strains were investigated for their H types by PCR amplification of fliC genes as described previously (37), and this was followed by restriction enzyme digestion of PCR products with HhaI and evaluation of restriction fragment length polymorphism patterns. Reference strains for H types H1 to H56 (47) were used as standard controls.
Detection of eae genes and of EAF plasmid- and bfpA-specific DNA sequences.
eae genes were detected by dot blot DNA hybridization as described previously (28). The 881-bp eae-specific DNA probe was derived from the STEC O157:H− strain E32511 (57) using the universal eae primers SK1 (5′ CCC GAA TTC GGC ACA AGC ATA AGC 3′) and SK2 (5′ CCC GGA TCC GTC TCG CCA GTA TTC G 3′) (48). The 397-bp EAF plasmid-specific gene probe used for DNA hybridization of Southern blotted plasmid DNA was derived by PCR from the E. coli K-12 strain MG1655 carrying the EAF plasmid pMAR7 (21). Labeling of the gene probes with digoxigenin-11-dUTP (Roche Diagnostics, Mannheim, Germany) and dot blot hybridization was performed as described previously (32). EAF plasmid-specific sequences were detected by PCR (21) as well as BFP (bfpA)-specific genes (27).
Subtyping of eae genes.
Typing of intimin genes into eae α, β, γ, and ɛ was performed by PCR using primer SK1 in combination with primers LP2 to LP5. Further subtyping of intimin genes was done by restriction fragment length polymorphism analysis of PstI-digested PCR products as described previously (48). PCRs for detection of new intimin variants ζ and η were developed on the basis of published nucleotide sequences (intimin ζ, accession no. AJ298279 and AJ271407; and intimin η, accession no. AJ308550) (67). Primers SK1 and LP6B (5′ TAG TTG TAC TCC CCT TAT CC 3′) were used to amplify a 2,430-bp intimin-ζ-specific product, and SK1 and LP8 (5′ TAG ATG ACG GTA AGC GAC 3′) were taken for amplification of the intimin η product (2,590 bp) (67). The PCR conditions were as follows: for intimin ζ, 30 cycles of 94°C for 30 s, 52°C for 60 s, and 72°C for 120 s; for intimin η, 30 cycles of 94°C for 30 s, 52°C for 60 s, and 72°C for 150 s. The reference strains used for identification of different intimin variants are listed elsewhere (67).
Detection of Stx1 and Stx2.
All E. coli isolates were investigated for cytotoxicity in the VERO cell toxicity assay; strains positive by this assay were examined for the production of Shiga toxin 1 (Stx1) and Stx2 with the VTEC-RPLA test (reverse passive latex agglutination test for detection of verocytotoxins VT1 and VT2; Denka-Seiken, Tokyo, Japan) as described previously (10). E. coli K-12 strains C600(H19) and C600(933W) were used as reference strains for Stx1 and Stx2 (8).
HEp-2 cell adhesion test.
The HEp-2 cell adhesion test was performed as described previously (8). Bacteria were allowed to absorb for 3 h or, alternatively, for 6 h before the cells were washed with phosphate-buffered saline (PBS), fixed with 70% methanol, and stained with 10% Giemsa (Merck). Adhesion tests were performed in the presence and absence of d-mannose (1%, wt/vol) in the growth medium (45). The following E. coli strains served as positive controls for different adhesion types: E. coli strains E20513 (O111:H2 eae+ eaf+ LA+) (9), DH5α(pSSS1) for DA (11), and 17-2 for enteroaggregative adherence (3). Strains E20518 (O128:H2, lacking eae and eaf) and HS served as negative controls for LA, DA, and enteroaggregative adherence, respectively (9).
FAS test.
The FAS test (34) was performed on HeLa cells seeded at 3 × 104 per well on eight-well chamber slides (Falcon, Becton Dickinson, Le Pont De Claix, France) and infected with EPEC strains cultivated overnight in Luria broth. The EPEC strains were inoculated at a multiplicity of infection of 100 on the HeLa cells incubated in Dulbecco's modified Eagle's medium (Gibco-Invitrogen Corporation, Karlsruhe, Germany) buffered with 25 mM HEPES (Gibco-Invitrogen) and supplemented with 5% fetal calf serum (Gibco-Invitrogen) and 1% d-mannose. After 4 to 5 h of interaction at 37°C in 5% CO2, monolayers were washed five times in PBS, fixed in 3% paraformaldehyde in PBS, and permeabilized for 5 min in 0.25% Triton in PBS. Then, the cells were stained with rhodamine phalloidin (Molecular Probes Europe, Leiden, The Netherlands) and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes Europe) to visualize F-actin accumulation at adhesion sites and adhering bacteria, respectively. Cells were examined by fluorescence microscopy with a DMRB light microscope (Leica Microsystems AG, Wetzlar, Germany). E. coli strain E2348/69 (O127:H6, eae+ eaf+ LA+) served as a positive control for the FAS test (31).
Conjugational transfer of EAF plasmid pMAR7, analysis of plasmid patterns, and detection of the pMAR7 plasmid by DNA hybridization with an EAF plasmid-specific gene probe.
The ampicillin transposon Tn801-tagged plasmid pMAR7 is a derivate of EAF plasmid pMAR2 from EPEC O127:H6 strain E2348/69 (2, 43). pMAR7 was transferred from its E. coli host strain HB101 (Lac−) by conjugation into the ampicillin-sensitive, Lac+ AEEC strains from infants and into the E. coli K-12 strain MG1655 (Lac+ Amps). Selection of transconjugants was made on minimal (M9 medium) agar plates supplemented with 0.2% lactose, 0.5% Casamino Acids (Difco), and ampicillin (25 μg/ml; Serva, Heidelberg, Germany). Transconjugant colonies were purified and tested for the presence of pMAR7 by EAF- and bfpA-specific PCR as described.
Plasmid DNA of AEEC and pMAR7-carrying derivatives was prepared with the Qiagen (Hilden, Germany) plasmid kit according to the instructions of the supplier. Plasmid DNA was separated on 0.7% agarose gels and Southern blotted as previously described (7). The Southern blot was hybridized with a digoxigenin-labeled EAF probe which was the EAF-specific PCR amplification product (21) from E. coli K-12 carrying plasmid pMAR7. HindIII digestion of plasmid DNA was performed according to the instructions of the supplier (Invitrogen GmbH, Karlsruhe, Germany).
RESULTS
Frequency and characterization of eae-positive E. coli (AEEC) from healthy infants.
A total of 482 E. coli strains were isolated from children in Berlin (205 infants and 264 strains) and Melbourne (184 infants and 218 strains). The 482 strains were examined for eae-specific gene sequences by dot blot DNA hybridization with the eae-specific gene probe (Materials and Methods). The presence of the eae gene in the probe-positive strains was confirmed by PCR using the universal eae-specific primers SK1 and SK2 (48). Fourteen (7.6%) of the Melbourne infants and 12 (5.9%) of the Berlin infants excreted AEEC. A total of 27 eae-positive strains from 26 children (12 girls and 14 boys) were isolated, and two serologically different AEEC (KK122/1 and KK122/9) were isolated from a stool sample of a 19-month-old boy from Berlin (Table 1). Only one (F88/6846-2, serotype O26:H11) of the 27 AEEC strains was verotoxic and produced Stx1. All other strains from infants did not show verotoxicity.
TABLE 1.
Properties of AEEC strains from healthy infants
| Strainj | Serotypeb | Intimin type | HEp-2 cell adherencec | FASd | pMAR7+a
|
|
|---|---|---|---|---|---|---|
| HEp-2 cell adherence | FAS | |||||
| LTEC94242 | 085:H49 | α1 | (DA) | − | (DA) | + |
| KK79/1 | O51:H49 | α1 | DA | + | NDe | ND |
| KK125/5 | ONT:[H6] | α2 | LAL | − | (DA) | +/− |
| KK130/1 | O132:[H34] | α2 | (DA) | − | LA | +/− |
| F88/6765-2 | O125:H6 | α2 | DA | − | LA | − |
| KK189/3 | O63:H6 | α2 | DA | − | ND | ND |
| F88/6846-2f | O26:H11 | β1 | DA | ++ | ND | ND |
| F89/8490-2 | O119:H2 | β1 | (DA) | − | LA | +++ |
| F91/2800-1 | Orough:H2 | β1 | LAL | + | LA | ++ |
| F91/3743-2 | O15:H2 | β1 | LAL | + | LA | + |
| LTEC93327 | O167:H9 | β1 | LAL | + | LA | +++ |
| LTEC94262 | O39:[H20] | β1 | DA | − | DA | − |
| KK111/1 | O26:[H11] | β1 | LAL | + | LA | +++ |
| KK83/10 | ONT:H6 | β2/δg | LAL | − | LA | +++ |
| KK122/9h | O88:[H5] | β2/δ | LAL | + | LA | +++ |
| KK141/10 | ONT:H6 | β2/δ | DA | − | ND | ND |
| KK145/10 | ONT:H6 | β2/δ | (DA) | − | (LA) | + |
| F89/9336-2 | O153:H11 | γ1 | LAL | − | LA | +++ |
| F90/3192-2 | O153:H11 | γ1 | DA | − | LA | + |
| LTEC93034 | O153:H11 | γ1 | DA | − | LA | ++ |
| KK72/5 | O156:H8 | γ1 | DA | ++ | LA | +++ |
| KK94/7 | O156:H8 | γ1 | (DA) | − | ND | ND |
| F91/1429-2 | O127:H45 | γ2 | (DA) | − | LA | − |
| KK77/1i | O76:H51 | γ2 | LA | +++ | LA | +++ |
| F91/0799-1 | O156:H1 | ζ | LAL | +/− | LA | + |
| LTEC94460 | O156:H1 | ζ | LAL | − | LA | ++ |
| KK122/1h | O2:H49 | η | DA | − | LA | +++ |
| MG1655 | E. coli K-12j | − | DA | − | DA | − |
pMAR7+, results obtained with pMAR7-carrying AEEC strains.
Strains for which an H type is indicated in brackets were either nonmotile (NM) or rough for their H antigen. The H type of these strains was detected by the RFLP typing of HhaI-digested amplified fliC genes as described previously (37).
Adherence patterns of AEEC to HEp-2 cells in the presence of 1% d-mannose. Interpretation of patterns was performed as described previously (55, 63). Abbreviations: DA, diffuse adherence; LA, localized adherence; LAL, localized adherence like; (DA) and (LA), weak adherence (≤10% of cells carried adhering bacteria)
Interpretation of FAS test results: +++, FAS positive reaction similar to that of the control strains E2348/69 (50 to 100% of adhering bacteria induced FAS response); ++, FAS positive (10 to 50% of adhering bacteria produced a FAS reaction); +, FAS positive (5 to 10% of adhering bacteria produced a FAS reaction); +/−, less than 5% of adhering bacteria caused a FAS-positive reaction; −, FAS negative or diffuse accumulation of actin.
ND, pMAR7 could not be introduced in these strains.
Positive for Stx1.
Strains KK122/9 and KK122/1 were isolated from the same child.
Positive for EAF-, and bfpA-specific DNA sequences.
E. coli strains whose designations begin with KK were isolated from children in Berlin; all others were from Melbourne infants.
The 27 AEEC strains belonged to 20 different O:H serotypes. Only four strains from four infants belonged to serotypes which are known to be associated with EPEC (O26:NM, O119:H2, O125:H6) and EHEC (O26:H11) groups (Table 1). The other 23 strains belonged to serotypes which do not belong to the reported groups of EPEC and EHEC (16; K. Bettelheim, unpublished data [www.sciencenet.com.au/vtectable.htm]). EAF- and bfpA-specific DNA sequences were detected in only one (KK77/1, serotype O76:H51) of the 27 AEEC strains.
Characterization of intimin genes and intimin subtypes.
Twenty-four of the 27 AEEC strains could be subtyped for their intimin genes by PCR and PstI restriction analysis of PCR products using primer SK1 in combination with primers LP2 to LP5 as described elsewhere (48). Three strains from infants (F91/0799-1, LTEC94460, and KK122/1) gave no product with these primer combinations although they were positive for eae by DNA hybridization and by PCR with universal eae primers SK1 and SK2. These strains could be characterized as carriers of the new variants intimin ζ (F91/0799-1 and LTEC94460, both serotype O156:H1) and intimin η (KK122/1, serotype O2:H49) (Table 1) using an extended PCR protocol for detection of intimin variants (67). Five intimin types, namely, α (6 strains), β (11 strains), γ (7 strains), ζ (2 strains), and η (one strain), were detected among the 27 AEEC strains, and none of the strains was positive for intimin types ɛ, ι, or κ (48, 67).
Construction of EAF plasmid-carrying AEEC derivative strains and analysis of plasmid patterns.
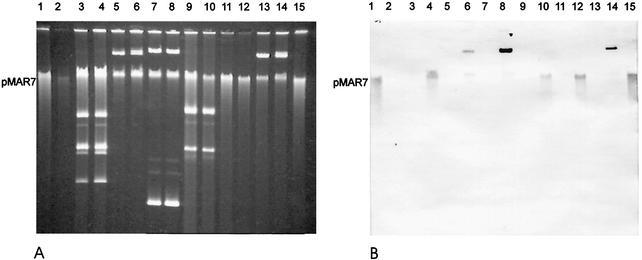
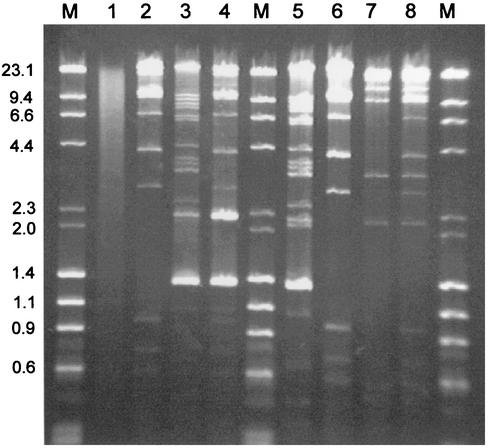
Since 26 of the 27 AEEC strains from infants were negative for an EAF plasmid and for the bfpA gene, we became interested in whether these strains could serve as hosts for the EAF plasmid pMAR7. Therefore, pMAR7 was introduced in the strains by conjugation, and 22 of the 27 AEEC strains yielded ampicillin-resistant transconjugants which stably inherited pMAR7 and tested positive by the bfpA and EAF-specific PCR (Table 1). The presence of pMAR7 in the transconjugant strains was shown by DNA hybridization of Southern blotted plasmid DNA with the EAF probe and by comparison of HindIII restriction patterns of plasmid DNA from parental AEEC and pMAR7-carrying derivative strains (Fig. 1 and 2, data not shown). Figure 1A shows a 0.7% agarose gel with separated plasmid DNA of representative AEEC strains and their pMAR7-carrying derivatives. A Southern blot of the gel shown in Fig. 1A was hybridized with the EAF probe under conditions of high stringency (Fig. 1B). The EAF probe hybridized only with plasmid DNA obtained from the pMAR7-carrying AEEC strains (Fig. 1B, lanes 1, 4, 6, 8, 10, 12, 14, and 15). Plasmid DNA preparations of some pMAR7-carrying strains yielded two hybridization signals with the EAF probe, one at the position of pMAR7 and one above it (Fig. 1, lanes 6, 8, and 14). This could be due to multimeric forms of the plasmids, which can be obtained with some strains using the Qiagen plasmid preparation kit (Qiagen plasmid purification handbook, September 2000). For further analysis of the plasmids present in AEEC and their pMAR7 derivatives we have digested these with HindIII and have compared their restriction fragment patterns on 0.7% agarose gels (Fig. 2). By this, pMAR7 characteristic restriction fragment patterns could be observed in all AEEC pMAR7 transconjugant strains (Fig. 2, lanes 2, 4, 6, and data not shown). In some strains the introduction of pMAR7 resulted in a loss of a resident plasmid which was present in the parental strain. Loss of plasmids was indicated by the absence of several HindIII restriction fragments in the pMAR7 derivative strain compared to the respective parental strain (Fig. 2, lanes 3, 4, 5, and 6). In contrast to plasmid DNA, chromosomal DNA yielded an indistinctive pattern when digested with HindIII, indicating that pMAR7 was present as an autonomous plasmid in the transconjugant E. coli K-12 and AEEC strains (Fig. 2, lane 1). All 22 transconjugant strains with pMAR7 were compared with the parental AEEC strains for HEp-2 cell adherence and in the FAS test (see below).
FIG. 1.
(A) Plasmid DNA patterns of AEEC and their pMAR7-carrying derivative strains separated on a 0.7% agarose gel. Strain MG1655 (E. coli K-12) served as the plasmid-free control strain. Lanes: 1, MG1655 (pMAR7); 2, MG1655; 3, LTEC93034; 4, LTEC93034 (pMAR7); 5, KK130/1; 6, KK130/1 (pMAR7); 7, KK122/9; 8, KK122/9 (pMAR7); 9, F91/1429-2; 10, F91/1429-2 (pMAR7); 11, F91/3743-2; 12, F91/3743-2 (pMAR7); 13, LTEC94460; 14, LTEC94460 (pMAR7); 15, MG1655 (pMAR7). (B) Southern blot of the agarose gel shown in panel A after hybridization with the EAF probe. A hybridization signal was obtained only with pMAR7 derivative strains. Multimeric forms of pMAR7 leading to two hybridization signals with the EAF probe were found in preparations from some of the strains (lanes 6, 8, and 14). The position of pMAR7 is indicated on both sides of the figure.
FIG. 2.
HindIII-digested plasmid DNA of AEEC and pMAR7-carrying derivative strains after gel electrophoresis on 1% agarose. Lanes: M, molecular weight standard (λ DNA/HindIII fragments and φX 174 RF DNA, HaeIII fragments (Invitrogen); 1, chromosomal DNA (MG1655); 2, MG1655 (pMAR7); 3, KK122/9; 4, KK122/9 (pMAR7); 5, KK125/5; 6, KK125/5 (pMAR7); 7, KK130/1; 8, KK130/1 (pMAR7).
Adherence of AEEC to HEp-2 cells.
All AEEC strains were investigated for their adherence to cultivated HEp-2 cells (Table 1). The adherence patterns were recorded as described previously (55, 63). Only the EAF-positive strain KK77/1 showed LA, whereas 16 strains showed DA, and 10 strains showed an LAL pattern. Six of the DA strains adhered only poorly (<10% of cells with adhering bacteria) to HEp-2 cells. Introduction of pMAR7 in the AEEC strains enhanced their adherence property and caused a change in the adherence pattern from DA or LAL to LA in 18 AEEC strains. Only three strains (LTEC94242, KK125/5, and LT94262) and the E. coli K-12 strain MG1655 (eae negative) were neither converted to an LA phenotype nor improved for their HEp-2 cell adherence by pMAR7 (Table 1). The LA profile of the strain KK77/1 was not changed after uptake of pMAR7. None of the 27 AEEC strains showed an enteroaggregative adherence pattern.
The adherence patterns of the AEEC strains were not altered by the presence or absence of d-mannose in the HEp-2 cell adherence assay medium. However, with some AEEC strains, fewer adhering bacteria and fewer HEp-2 cells with attached bacteria were found when 1% d-mannose was present in the test medium.
FAS response with AEEC from infants.
The 27 AEEC strains were investigated for their AE phenotype by the FAS test (34, 35). Accumulation of actin under the bacteria was observed with 10 AEEC strains; 7 of these were weakly FAS positive (≤10% of adhering bacteria produced FAS), and only 3 strains (KK77/1, KK72/5, and F88/6846-2) showed a strong FAS response (10 to 100% of adhering bacteria were FAS positive), which was comparable to the positive control strain E2348/69 (Table 1). All other AEEC strains were clearly FAS negative.
By introduction of pMAR7, 17 of the AEEC strains became FAS positive or showed a stronger FAS response than the parental strains. Nine pMAR7+ transconjugant strains showed a FAS reaction comparable to that of the FAS-positive control strain E2348/69 (Table 1). Five AEEC (pMAR7+) strains showed a weak positive (KK125/5 and KK130/1) or negative (F88/6765-2, LTEC94262, and F91/1429-2) FAS response (Table 1). Among the five strains, two, LTEC94262 and KK125/5, were adhered poorly to HEp-2 cells and did not show an LA phenotype even when carrying pMAR7.
By comparing the FAS reactions of strains with different intimin types we found that all six AEEC strains with intimin α were FAS negative (n = 5) or only weakly FAS positive (n = 1). The introduction of pMAR7 did not result in an increase of the FAS reaction with these strains (Table 1). In contrast, the FAS positivity of 11 out of 18 strains carrying either intimin β, γ, ζ, or η were significantly enhanced (10 to 100% of adhering bacteria produced FAS) when these strains were carrying pMAR7.
DISCUSSION
E. coli strains encoding intimin and other functions needed for the AE phenotype (AEEC strains) were found to be heterogeneous according to their virulence markers and human pathogenicity. Two subgroups of AEEC, called EPEC and EHEC, are recognized as important human pathogens causing diarrhea (EPEC and EHEC) and hemolytic-uremic syndrome (EHEC). Typical EPEC strains harbor an EAF plasmid which encodes functions needed for intestinal colonization and expression of the AE phenotype, whereas EHEC strains are characterized by production of Shiga toxins (42, 63). A third group of AEEC strains, called atypical EPEC strains, is formed by strains which harbor the LEE but which are negative for production of Stx and the EAF plasmid (42, 63). Atypical EPEC strains were isolated from feces of healthy and diarrheic humans, but in contrast to typical EPEC or EHEC, their role as pathogens is less clear (20, 25, 36, 42, 56).
In this work, we were interested in the occurrence and properties of AEEC strains in fecal samples from children without enteric disease in developed countries of the Northern and Southern Hemispheres. AEEC strains were isolated at similar rates from Berlin (5.9%) and Melbourne (7.6%) infants, indicating that these strains are naturally occurring in the stool flora of healthy children in different parts of the world (20, 25, 36). Only 1 of the 27 AEEC isolates from healthy children was identified as EHEC (F88-6846-2 Stx1+), and a second AEEC strain (KK77/1) carried an EAF plasmid. The remaining 25 AEEC strains were negative for Stx, bfpA, and EAF and were thus classified as atypical EPEC (42, 63).
Only three of the atypical EPEC strains from children (KK111/1, F89/8490-2, and F88/6765-2) belonged to serotypes (O26:H11, O119:H2, and O125:H6) which are frequently associated with diarrhea in infants (63). The other 22 atypical EPEC strains from healthy children belonged either to serotypes which were rarely (O15:H2, O39:NM,O63:H6, and O156:H8) or not associated with diarrheal disease (12, 29, 65, 66). In order to obtain more information on the potential virulence properties of these AEEC strains for infants, we have investigated the HEp-2 cell adherence and the AE phenotype by the FAS assay. LA to HEp-2 cells was only found with EAF-positive AEEC strain KK77/1. All other AEEC strains from children showed DA or LAL adherence patterns, which is commonly found with atypical EPEC strains (55, 63). The number of bacteria adhering to HEp-2 cells was lower with some AEEC strains when d-mannose was present in the assay medium. This could indicate that type 1 pili play a role in adherence of these strains but also points to catabolite repression of adhesion, as was described for some EHEC strains (45).
When tested for the AE phenotype, only 3 of the 27 AEEC strains showed a FAS response which was similar to that of the FAS-positive control strain E2348/69. These three strains were KK77/1 (typical EPEC), F88-6846-2 (EHEC), and 1 of the 25 atypical EPEC strains (KK72/5). The remaining atypical EPEC strains were either FAS negative or gave only a weak FAS-positive reaction (Table 1). In order to investigate if the weak or negative FAS reaction found in these strains was due to poor adherence to eukaryotic cells or a defect in the genes needed for the AE phenotype, we have introduced the EAF plasmid pMAR7 into the AEEC strains. This plasmid carries functions promoting both colonization (bfp operon) and AE property (per/bfpTVW) of the strains (17, 26, 41). The introduction of pMAR7 had a positive effect on HEp-2 cell adherence for most of the AEEC strains and converted 11 FAS-negative AEEC strains to FAS positive and increased the FAS reaction of six FAS-positive AEEC strains. These results show that the genes required for the AE phenotype are functional in these 17 atypical EPEC strains and that the weak or negative FAS response of the wild-type AEEC strains could be due to weak adhesion and/or weak expression of the LEE-encoded genes which is compensated by the functions encoded by pMAR7 (Table 1). In contrast to that, five atypical EPEC strains showed different responses when carrying pMAR7. Two of these (LTEC94242 and KK125/5) were not converted to LA but showed an improved FAS response, whereas two other strains (F88-6765-2 and F91/1429-2) became LA positive but remained FAS negative. The strain LTEC94262 remained negative for both the FAS reaction and LA. These results indicate that functions needed for colonization (LTEC94242, KK125/5 and LTEC94262) and upregulation of LEE genes (F88-6765-2, F91-1429-2, and LTEC94262) are not well expressed in these strains. A number of chromosomal genes encoding functions involved in adherence of EPEC and EHEC and in regulation of LEE gene expression were recently described (13, 44, 59, 62). These include the product of the efa1 gene, which contributes to an adhesive phenotype in EHEC (44), and the trcA product, which is needed for the assembly of BFP and EPEC colonization (62). efa1 and trcA genes are genetically not linked to the LEE, although the presence of these genes was found to be closely associated with E. coli strains carrying LEE genes such as EPEC and EHEC (44, 62). Other chromosomally encoded genes such as gadX were found to be involved in regulation of perA expression and expression of EPEC virulence genes (59).
Furthermore, it is also possible that the FAS-negative strains F88/6765-2, F91/1429-2, and LTEC94262 are defective in the LEE genes required for production of the AE lesion or in the ler gene product which antagonizes H-NS-dependent repression of LEE genes (13). It was also shown that E. coli carrying the LEE can lack some of the LEE genes, which results in an AE-negative phenotype (53). Further work is needed to explore the basis of the LA- and FAS-negative phenotype found in some AEEC strains from our study.
Five of the 27 AEEC strains (KK79/1, KK189/3, F88/6846-2, KK141/10, and KK94/7) did not yield Ampr transconjugant colonies after repeated matings with the donor strain HB101 (pMAR7+). We do not know if this is due to incompatibility between pMAR7 and resident plasmids present in these strains or to other factors affecting the entry or replication of pMAR7 in the AEEC recipients. Most of the AEEC strains from children were found to carry large plasmids, and in some strains the resident plasmids became lost when selection was made for pMAR7 (Fig. 2 and data not shown). The role of the resident plasmids of the AEEC strains is not known but it appears possible that some of these share incompatibility with EAF plasmids and present therefore a natural barrier against incoming IncF plasmids such pMAR7 and other plasmids of the EAF family (39, 42, 61).
Subtyping of the eae genes present in the 27 AEEC strains from infants revealed that 24 of these carried either intimin α, β, or γ. These three intimin variants are most frequent in EPEC and EHEC strains which were isolated from human patients (1, 48, 67). Three of the atypical EPEC strains (F91/0799-1, LTEC94460, and KK122/1) showed the new described intimin types ζ and η. These intimin variants were associated with a few serotypes of E. coli which do not belong to typical EPEC or EHEC groups (67). These new intimin variants were found to be functionally expressed in their host, since all three strains with the new intimin variants became FAS positive when carrying pMAR7.
Apart from our study, AEEC strains have been isolated at a similar rate from healthy infants in France (6.5%) (20), Brazil (7.7%) (25), and Chile (36). Most of the strains from these studies were negative for the EAF plasmid and for Stx, suggesting atypical EPEC. It is not clear if these isolates represent human pathogens since they were not investigated for expression of LEE genes and for their AE phenotype in the FAS assay (20, 25).
Among human patients, children younger than 2 years of age are primarily affected by EPEC infections (42). All children investigated in our study belonged to this age-related risk group. In order to determine the pathogenic potential of their AEEC isolates, we have investigated the strains for their AE phenotype by the FAS assay. Our findings indicate that most of the AEEC strains from these infants either do not or only weakly express an AE phenotype which could explain why these strains did not cause diarrheal disease. However, most of the AEEC strains from healthy infants were not affected in the function of LEE-associated genes because they became clearly AE positive after uptake of pMAR7. It has been reported that the degree of actin aggregation is lower with AEEC strains adhering in a diffuse manner to eukaryotic cells (4, 14). Therefore, the weak expression of the AE phenotype in the AEEC wild-type strains might be related to their adhesion properties. A similar observation was made in another study showing that the introduction of a plasmid encoding the F1845 adhesin improved both the adhesion and FAS reaction in FAS-negative and poorly adhering AEEC strains (14).
It is possible that atypical AEEC strains which are not or are only weakly FAS positive play a role as natural immunizing strains to intimin and other proteins encoded by the LEE which induce antibody production in infected humans (30, 42). On the other hand, our finding that the EAF plasmid could be introduced and that its virulence functions were expressed in most of the atypical AEEC strains points also to their role as a reservoir for new emerging human pathogenic EPEC strains. Further studies are needed to explore the reservoir of atypical AEEC in humans and animals, their relationship to known pathogenic EPEC and EHEC strains, and their role for the host as part of the microecology of the human intestinal tract.
Acknowledgments
L. Beutin, K. Gleier, E. Oswald, H. Schmidt, and S. Zimmermann were supported by funds from the European Commission project Attaching and Effacing Escherichia coli Infections (reference no. QLK2-CT-2000-00600).
We thank J. L. Pearce and R. K. L. Luke for assistance with the initial collection and serotyping of the strains of the Melbourne collection.
Editor: V. J. DiRita
REFERENCES
- 1.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldini, M. M., J. B. Kaper, M. M. Levine, D. C. Candy, and H. W. Moon. 1983. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J. Pediatr. Gastroenterol. Nutr. 2:534-538. [DOI] [PubMed] [Google Scholar]
- 3.Baudry, B., S. J. Savarino, P. Vial, J. B. Kaper, and M. M. Levine. 1990. A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen. J. Infect. Dis. 161:1249-1251. [DOI] [PubMed] [Google Scholar]
- 4.Beinke, C., S. Laarmann, C. Wachter, H. Karch, L. Greune, and M. A. Schmidt. 1998. Diffusely adhering Escherichia coli strains induce attaching and effacing phenotypes and secrete homologs of Esp proteins. Infect. Immun. 66:528-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettelheim, K. A., L. Beutin, K. Gleier, J. L. Pearce, R. K. J. Luke, and S. Zimmermann. 2003. Serotypes of Escherichia coli isolated from healthy children in Berlin, Germany and Melbourne, Australia. Comp. Immunol. Microbiol. Infect. Dis. 26:55-63. [DOI] [PubMed] [Google Scholar]
- 6.Bettelheim, K. A., and C. J. Thompson. 1987. New method of serotyping Escherichia coli: implementation and verification. J. Clin. Microbiol. 25:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutin, L., M. Bulte, A. Weber, S. Zimmermann, and K. Gleier. 2000. Investigation of human infections with verocytotoxin-producing strains of Escherichia coli (VTEC) belonging to serogroup O118 with evidence for zoonotic transmission. Epidemiol. Infect. 125:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutin, L., D. Geier, S. Zimmermann, and H. Karch. 1995. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J. Clin. Microbiol. 33:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beutin, L., I. Orskov, F. Orskov, S. Zimmermann, J. Prada, H. Gelderblom, R. Stephan, and T. S. Whittam. 1990. Clonal diversity and virulence factors in strains of Escherichia coli of the classic enteropathogenic serogroup O114. J. Infect. Dis. 162:1329-1334. [DOI] [PubMed] [Google Scholar]
- 10.Beutin, L., S. Zimmermann, and K. Gleier. 1996. Rapid detection and isolation of Shiga-like toxin (verocytotoxin)-producing Escherichia coli by direct testing of individual enterohemolytic colonies from washed sheep blood agar plates in the VTEC-RPLA assay. J. Clin. Microbiol. 34:2812-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilge, S. S., C. R. Clausen, W. Lau, and S. L. Moseley. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J. Bacteriol. 171:4281-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bokete, T. N., T. S. Whittam, R. A. Wilson, C. R. Clausen, C. M. O'Callahan, S. L. Moseley, T. R. Fritsche, and P. I. Tarr. 1997. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J. Infect. Dis. 175:1382-1389. [DOI] [PubMed] [Google Scholar]
- 13.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 14.Cantey, J. R., and S. L. Moseley. 1991. HeLa cell adherence, actin aggregation, and invasion by nonenteropathogenic Escherichia coli possessing the eae gene. Infect. Immun. 59:3924-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.China, B., F. Goffaux, V. Pirson, and J. Mainil. 1999. Comparison of eae, tir, espA and espB genes of bovine and human attaching and effacing Escherichia coli by multiplex polymerase chain reaction. FEMS Microbiol. Lett. 178:177-182. [DOI] [PubMed] [Google Scholar]
- 16.Donnenberg, M. S. 1995. Enteropathogenic Escherichia coli, p. 709-726. In M. J. Blaser et al. (ed.), Infections of the gastrointestinal tract. Raven Press, New York, N.Y.
- 17.Donnenberg, M. S., J. A. Giron, J. P. Nataro, and J. B. Kaper. 1992. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol. Microbiol. 6:3427-3437. [DOI] [PubMed] [Google Scholar]
- 18.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 19.Fitzhenry, R. J., S. Reece, L. R. Trabulsi, R. Heuschkel, S. Murch, M. Thomson, G. Frankel, and A. D. Phillips. 2002. Tissue tropism of enteropathogenic Escherichia coli strains belonging to the O55 serogroup. Infect. Immun. 70:4362-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forestier, C., M. Meyer, S. Favre-Bonte, C. Rich, G. Malpuech, C. Le Bouguenec, J. Sirot, B. Joly, and C. De Champs. 1996. Enteroadherent Escherichia coli and diarrhea in children: a prospective case-control study. J. Clin. Microbiol. 34:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franke, J., S. Franke, H. Schmidt, A. Schwarzkopf, L. H. Wieler, G. Baljer, L. Beutin, and H. Karch. 1994. Nucleotide sequence analysis of enteropathogenic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J. Clin. Microbiol. 32:2460-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 23.Giron, J. A., M. S. Donnenberg, W. C. Martin, K. G. Jarvis, and J. B. Kaper. 1993. Distribution of the bundle-forming pilus structural gene (bfpA) among enteropathogenic Escherichia coli. J. Infect. Dis. 168:1037-1041. [DOI] [PubMed] [Google Scholar]
- 24.Giron, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 25.Gomes, T. A., P. M. Griffin, C. Ivey, L. R. Trabulsi, and S. R. Ramos. 1996. EPEC infections in Sao Paulo. Rev. Microbiol. 27:25-33. [Google Scholar]
- 26.Gomez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunzburg, S. T., N. G. Tornieporth, and L. W. Riley. 1995. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J. Clin. Microbiol. 33:1375-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammermueller, J., S. Kruth, J. Prescott, and C. Gyles. 1995. Detection of toxin genes in Escherichia coli isolated from normal dogs and dogs with diarrhea. Can. J. Vet. Res. 59:265-270. [PMC free article] [PubMed] [Google Scholar]
- 29.Hedberg, C. W., S. J. Savarino, J. M. Besser, C. J. Paulus, V. M. Thelen, L. J. Myers, D. N. Cameron, T. J. Barrett, J. B. Kaper, and M. T. Osterholm. 1997. An outbreak of foodborne illness caused by Escherichia coli O39:NM, an agent not fitting into the existing scheme for classifying diarrheogenic E. coli. J. Infect. Dis. 176:1625-1628. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins, C., H. Chart, H. R. Smith, E. L. Hartland, M. Batchelor, R. M. Delahay, G. Dougan, and G. Frankel. 2000. Antibody response of patients infected with verocytotoxin-producing Escherichia coli to protein antigens encoded on the LEE locus. J. Med. Microbiol. 49:97-101. [DOI] [PubMed] [Google Scholar]
- 31.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaddu-Mulindw, D. H., T. Aisu, K. Gleier, S. Zimmermann, and L. Beutin. 2001. Occurrence of Shiga toxin-producing Escherichia coli in fecal samples from children with diarrhea and from healthy zebu cattle in Uganda. Int. J. Food Microbiol. 66:95-101. [DOI] [PubMed] [Google Scholar]
- 33.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 34.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knutton, S., A. D. Phillips, H. R. Smith, R. J. Gross, R. Shaw, P. Watson, and E. Price. 1991. Screening for enteropathogenic Escherichia coli in infants with diarrhea by the fluorescent-actin staining test. Infect. Immun. 59:365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine, M. M., V. Prado, H. Lior, and R. Lagos. 1996. Epidemiologic studies of diarrhea associated with enteropathogenic Escherichia coli (EPEC) in Santiago, Chile. Rev. Microbiol. 27:40-44. [Google Scholar]
- 37.Machado, J., F. Grimont, and P. A. Grimont. 2000. Identification of Escherichia coli flagellar types by restriction of the amplified fliC gene. Res. Microbiol. 151:535-546. [DOI] [PubMed] [Google Scholar]
- 38.Marches, O., J. P. Nougayrede, S. Boullier, J. Mainil, G. Charlier, I. Raymond, P. Pohl, M. Boury, J. De Rycke, A. Milon, and E. Oswald. 2000. Role of Tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect. Immun. 68:2171-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McConnell, M. M., H. Chart, S. M. Scotland, H. R. Smith, G. A. Willshaw, and B. Rowe. 1989. Properties of adherence factor plasmids of enteropathogenic Escherichia coli and the effect of host strain on expression of adherence to HEp-2 cells. J. Gen. Microbiol. 135:1123-1134. [DOI] [PubMed] [Google Scholar]
- 40.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 42.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nataro, J. P., K. O. Maher, P. Mackie, and J. B. Kaper. 1987. Characterization of plasmids encoding the adherence factor of enteropathogenic Escherichia coli. Infect. Immun. 55:2370-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 45.Nishikawa, Y., S. M. Scotland, H. R. Smith, G. A. Willshaw, and B. Rowe. 1995. Catabolite repression of the adhesion of Vero cytotoxin-producing Escherichia coli of serogroups 0157 and 0111. Microb. Pathog. 18:223-229. [DOI] [PubMed] [Google Scholar]
- 46.Okeke, I. N., J. A. Borneman, S. Shin, J. L. Mellies, L. E. Quinn, and J. B. Kaper. 2001. Comparative sequence analysis of the plasmid-encoded regulator of enteropathogenic Escherichia coli strains. Infect. Immun. 69:5553-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orskov, F., and I. Orskov. 1984. Serotyping of Escherichia coli. Methods Microbiol. 14:43-112. [Google Scholar]
- 48.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paton, A. W., P. A. Manning, M. C. Woodrow, and J. C. Paton. 1998. Translocated intimin receptors (Tir) of Shiga-toxigenic Escherichia coli isolates belonging to serogroups O26, O111, and O157 react with sera from patients with hemolytic-uremic syndrome and exhibit marked sequence heterogeneity. Infect. Immun. 66:5580-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pelayo, J. S., I. C. Scaletsky, M. Z. Pedroso, V. Sperandio, J. A. Giron, G. Frankel, and L. R. Trabulsi. 1999. Virulence properties of atypical EPEC strains. J. Med. Microbiol. 48:41-49. [DOI] [PubMed] [Google Scholar]
- 51.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reece, S., C. P. Simmons, R. J. Fitzhenry, S. Matthews, A. D. Phillips, G. Dougan, and G. Frankel. 2001. Site-directed mutagenesis of intimin alpha modulates intimin-mediated tissue tropism and host specificity. Mol. Microbiol. 40:86-98. [DOI] [PubMed] [Google Scholar]
- 53.Sandner, L., L. E. Eguiarte, A. Navarro, A. Cravioto, and V. Souza. 2001. The elements of the locus of enterocyte effacement in human and wild mammal isolates of Escherichia coli: evolution by assemblage or disruption? Microbiology 147:3149-3158. [DOI] [PubMed] [Google Scholar]
- 54.Scaletsky, I. C., S. H. Fabbricotti, K. R. Aranda, M. B. Morais, and U. Fagundes-Neto. 2002. Comparison of DNA hybridization and PCR assays for detection of putative pathogenic enteroadherent Escherichia coli. J. Clin. Microbiol. 40:1254-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scaletsky, I. C., J. S. Pelayo, R. Giraldi, J. Rodrigues, M. Z. Pedroso, and L. R. Trabulsi. 1996. EPEC adherence to HEp-2 cells. Rev. Microbiol. 27:58-62. [Google Scholar]
- 56.Schmidt, H., H. Russmann, A. Schwarzkopf, S. Aleksic, J. Heesemann, and H. Karch. 1994. Prevalence of attaching and effacing Escherichia coli in stool samples from patients and controls. Zentbl. Bakteriol. 281:201-213. [DOI] [PubMed] [Google Scholar]
- 57.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H- strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scotland, S. M., G. A. Willshaw, H. R. Smith, and B. Rowe. 1990. Properties of strains of Escherichia coli O26:H11 in relation to their enteropathogenic or enterohemorrhagic classification. J. Infect. Dis. 162:1069-1074. [DOI] [PubMed] [Google Scholar]
- 59.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 60.Tarr, C. L., and T. S. Whittam. 2002. Molecular evolution of the intimin gene in O111 clones of pathogenic Escherichia coli. J. Bacteriol. 184:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 62.Tobe, T., I. Tatsuno, E. Katayama, C. Y. Wu, G. K. Schoolnik, and C. Sasakawa. 1999. A novel chromosomal locus of enteropathogenic Escherichia coli (EPEC), which encodes a bfpT-regulated chaperone-like protein, TrcA, involved in microcolony formation by EPEC. Mol. Microbiol. 33:741-752. [DOI] [PubMed] [Google Scholar]
- 63.Trabulsi, L. R., R. Keller, and G. T. Tardelli. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vallance, B. A., and B. B. Finlay. 2000. Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8799-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vieira, M. A., J. R. Andrade, L. R. Trabulsi, A. C. Rosa, A. M. Dias, S. R. Ramos, G. Frankel, and T. A. Gomes. 2001. Phenotypic and genotypic characteristics of Escherichia coli strains of non-enteropathogenic E. coli (EPEC) serogroups that carry EAE and lack the EPEC adherence factor and Shiga toxin DNA probe sequences. J. Infect. Dis. 183:762-772. [DOI] [PubMed] [Google Scholar]
- 66.Yamazaki, M., M. Saito, K. Inuzuka, S. Shima, H. Taniwaki, and K. Ito. 1997. eaeA genes in Escherichia coli derived from Japanese patients with sporadic diarrhea. Kansenshogaku Zasshi 71:1059-1065. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, W. L., B. Kohler, E. Oswald, L. Beutin, H. Karch, S. Morabito, A. Caprioli, S. Suerbaum, and H. Schmidt. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu, C., T. S. Agin, S. J. Elliott, L. A. Johnson, T. E. Thate, J. B. Kaper, and E. C. Boedeker. 2001. Complete nucleotide sequence and analysis of the locus of enterocyte effacement from rabbit diarrheagenic Escherichia coli RDEC-1. Infect. Immun. 69:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]