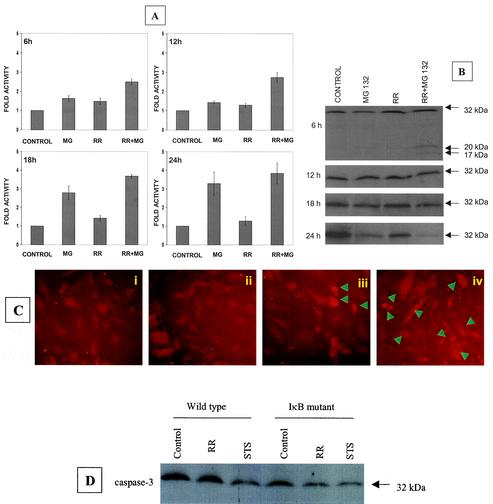
FIG. 1.
Activation of caspase-3 during R. rickettsii-induced apoptosis. EC were infected with R. rickettsii (≈5 × 104 PFU/cm2) for 2 h, after which extracellular bacteria were removed by washing and the cells were incubated further as indicated. NF-κB activation was attenuated by incubating with MG132 (25 μM), an inhibitor of proteasome function, for 30 min prior to and during the infection. (A) Analysis of caspase-3 activity in EC extracts, with DEVD-pNA as the chromogenic substrate. EC were left untreated (control), treated with MG132 (MG), or infected in the absence (RR) or presence (RR + MG) of MG132. Measurements of caspase activity were performed at 6 h, 12 h, 18 h, and 24 h. The baseline values (mean ± SEM) for untreated-uninfected cells at these times were 0.010 ± 0.001, 0.018 ± 0.003, 0.022 ± 0.004, and 0.025 ± 0.005 absorbance units, respectively. For comparison, the activity level in controls was assigned a value of 1, and the results are presented as mean ± SEM from three independent experiments. (B) Western blot analysis of caspase-3 at different times postinfection. (C) Indirect immunofluorescent staining for detection of cleaved caspase-3 at 12 h. The conditions were (i) uninfected EC, (ii) EC exposed to MG132, (iii) EC infected with R. rickettsii, and (iv) EC infected with R. rickettsii in the presence of MG132. Arrowheads indicate cells that are positive for the cleaved caspase-3 fragment and exhibit morphologic features of apoptosis. (D) Western blot analysis of cell extracts from wild-type (empty LXSN vector) and IκBα mutant-expressing T-24 cells, showing processing of procaspase-3 after 6 h of infection. Exposure of cells to staurosporine (STS) was included as a positive control. Equal amounts of protein were resolved on 12% polyacrylamide gels, blotted to a nitrocellulose membrane, probed with an anticaspase-3 antibody, and visualized by chemiluminescent detection.