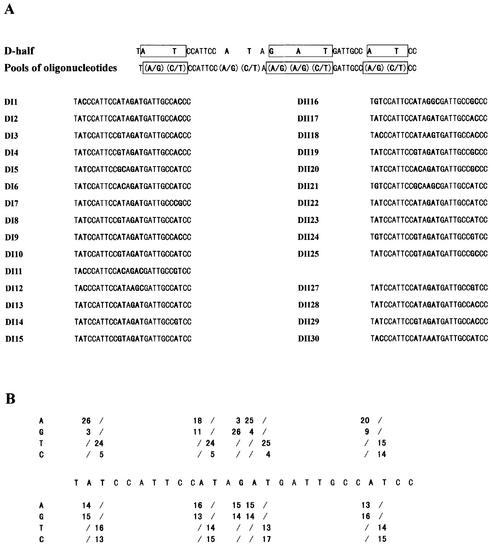
Figure 5.
NodD-binding sites selected from pools of oligonucleotides carrying nine mutated nucleotides in the context of the D-half sequence. (A) Sequences of 29 selected probes bound specifically by NodD. The wild-type D-half sequence context is given in the first line, with the consensus conserved-sequence boxed. The mutated nucleotides are in bold: nucleotide A was randomly mutated to A or G, nucleotide T was mutated to T or C, and nucleotide G to A or G in a 1:1 ratio. Pools of 512 species of mutant oligonucleotides are denoted in the second line. The oligonucleotides selected from two independent selections (DI and DII) were aligned, with the mutant nucleotides in bold. (B) Summary of frequencies of nucleotide residues for selected (top bases) and unselected (bottom bases) oligonucleotides. The nucleotides of the consensus AT-N10-GAT-N7-A are clearly biased for NodD binding. Even the ‘control’ nucleotides A and T of the non-conserved consensus sequence are also biased. Out of 29 selected oligonucleotides, there are 21 with the sequence AT-N10-GAT selected.