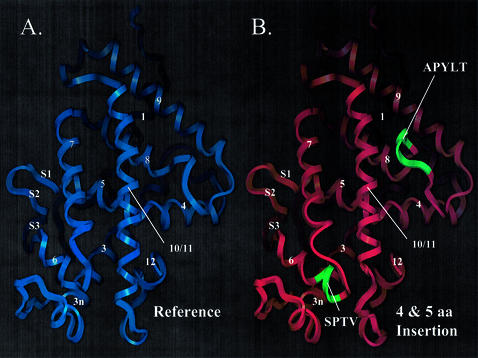
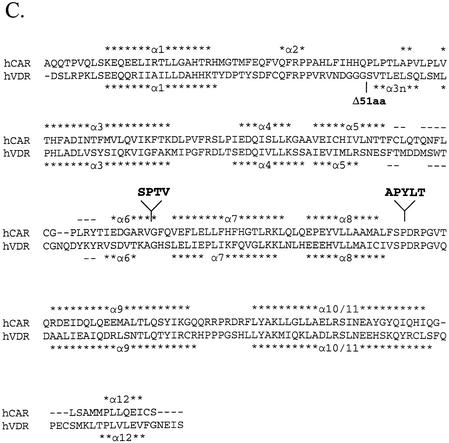
Figure 5.
hCAR structure model suggests that hCAR isoforms possess altered ligand-binding domain structure. (A) hCAR-REF was modeled using the hVDR X-ray crystallographic structure [PDB accession code 1DB1(31)] as a template. α-Helices are labeled 1–12. β-Strands are labeled S1, 2 or 3, according to the VDR structure. (B) hCAR-dblINS was generated using the same procedure as for the hCAR-REF model. Secondary structure is labeled as in the reference model. Inserted amino acid sequences are shown in green and are labeled with their corresponding amino acid sequences. The 4 amino acid insertion (SPTV) is predicted to extend α-helix 6 in the C-terminal direction. The 5 amino acid insertion (APYLT) is predicted to extend the loop between α-helices 8 and 9. (C) hCAR (upper sequence) was aligned with the sequence of hVDR (lower sequence) used in the generation of the 1DB1 structure. α-Helices are indicated by asterisks, and β-strands are indicated by dashes. The locations of amino acid insertions are indicated by arrows and shown as text in bold font above the sequences. The location of the 51 amino acid deletion in the VDR model is also indicated.