Abstract
Sex determination in the hermaphrodite germ line of Caenorhabditis elegans is controlled posttranscriptionally. The switch from spermatogenesis to oogenesis relies on regulation of the fem-3 sex-determining gene via a regulatory element in the fem-3 3′ untranslated region. Previous work showed that at least six mog genes are required for repression by the fem-3 3′ untranslated region, and that one of those genes, mog-1, encodes a DEAH-box protein. In this paper, we report the cloning of mog-4 and mog-5 and the finding that mog-4 and mog-5 also encode DEAH-box proteins. Our molecular identification of mog-4 and mog-5 relied on genetic mapping and transformation rescue and was confirmed by a missense mutation in each gene. A phylogenetic analysis revealed that the C. elegans MOG-1, MOG-4, and MOG-5 proteins are closely related to the yeast proteins PRP16, PRP2, and PRP22, respectively. In view of their effect on fem-3 regulation and their homology to PRP16, PRP2, and PRP22, we propose that MOG-1, MOG-4, and MOG-5 are required for posttranscriptional regulation, perhaps by modifying the conformation of ribonucleoprotein complexes.
In Caenorhabditis elegans, the hermaphrodite produces sperm during the fourth larval stage and oocytes in the adult (1). Normally, the fem-3 sex-determining gene promotes male fates transiently in the XX hermaphrodite germ line (2). In dominant regulatory fem-3 mutants, the XX germ line is masculinized: sperm are made continuously and no oogenesis occurs (3). Molecular analyses of these fem-3 gain-of-function (gf) mutations revealed the point mutation element (PME), a cis-acting regulatory element in the fem-3 3′ untranslated region (UTR), that is required for the switch from spermatogenesis to oogenesis (4). Furthermore, the fem-3 3′ UTR is sufficient to repress a reporter transgene and this repression is PME-dependent (5). These findings have led to a model in which fem-3 is repressed posttranscriptionally to achieve the hermaphrodite switch from spermatogenesis to oogenesis in the XX germ line.
Six mog genes (mog-1–mog-6) are key regulators of the hermaphrodite sperm/oocyte switch and are also critical for PME-mediated repression in reporter assays (5–7). XX animals homozygous for mutations in any of these mog genes fail to switch from spermatogenesis to oogenesis. In addition, the mog genes are required maternally for embryogenesis. It is not yet clear whether the mog gene products act directly or indirectly to promote PME-repression. One clue is that mog-1 encodes a member of the DEAH box protein family, suggesting that it acts at a posttranscriptional level (8).
In this study, we report that mog-4 and mog-5 encode proteins of the DEAH-box family. Because three of the six mog genes encode members of this same family (ref. 8 and this work), we explored all DEAH-box proteins in the C. elegans genome by phylogenetic analysis and by RNA-mediated interference (RNAi). We found that mog-1, mog-4, and mog-5 are closely related to the yeast splicing factors PRP16, PRP2, and PRP22, respectively, and that they are the only mog genes that encode DEAH-box proteins.
Materials and Methods
Cloning mog-4.
Initial mapping placed mog-4 to the left of unc-52 on chromosome II (6). C04H5.6, which encodes a DEAH-box protein, resides in this region. Rescue was attempted with C04H5 or a 12.8-kb XbaI–BstXI subclone (pAP1) covering C04H5.6. DNA injected was 5 ng/μl of the C04H5 cosmid or pAP1, 30 ng/μl pRF4 roller DNA, and 70 ng/μl Haemophilus influenzae genomic DNA as carrier. The H. influenzae DNA was added to create a complex extrachromosomal array, which facilitates germline expression (9). Adults injected were unc-4(e120) mog-4(q233)/mnC1. The penetrance of mog-4(q233) was 100% at 20°C: all adults scored were Mog: no oocytes and a large excess of sperm (n = >100 worms scored). Unc-4 progeny carrying either C04H5 or pAP1 as transgenes were scored for fertility; when fertile, such animals had low brood sizes and segregated many dead embryos, as expected for a weakly rescued mog-4 mutant. From 29 worms injected, 86 F1 rollers were obtained. Among those rollers were seven sterile UncMog worms and three fertile Uncs, which were rescued. One fertile line was maintained for several generations. To confirm the identity of mog-4 as C04H5.6, we sequenced the corresponding genomic DNA from mog-4(q233) mutants: genomic DNA was extracted from mog-4 homozygotes and C04H5.6 amplified by PCR using Expand Taq DNA polymerase (Boehringer Mannheim). PCR products were sequenced directly and compared with published sequence.
Cloning mog-5.
mog-5 maps between unc-85 and dpy-10 (ref. 6 and this study); it is deleted by mnDf4, but not by mnDf96 or mnDf30. EEED8.5, which was predicted to encode a DEAH-protein, resides in this region. Rescue was attempted with cosmid EEED8, pJK610 (a 7.4-kb HindIII subclone covering EEED8.5), or pJK611, a variant of pJK610 with a 2,344-nt deletion (a BspEI fragment). The composition of injected DNAs was similar to that described for mog-4, except with relevant DNAs. Adults injected were mog-5(q449) dpy-10(e128)/unc-85(e1414); progeny were scored for fertile Dpy-10. On injection of 22 heterozygotes, 41 fertile Dpy were obtained of which 16 were rescued mog-5 homozygotes and 25 were recombinants. Rescued mutants were distinguished from recombinants by their reduced fertility (3–100 eggs laid per individual) and reduced embryonic viability. A few stable rescued lines were obtained. Rescue (the number of fertile rolling Uncs/total rolling Uncs) was ≈80% for mog-5, but <35% for mog-4, even though similar injection mixtures were used.
Isolation of cDNAs.
C. elegans cDNA libraries (λRB1 and λRB2, provided by R. Barstead, Oklahoma Medical Research Foundation, Oklahoma City, OK) were screened with genomic mog-4 and mog-5 probes. To obtain 5′ ends, we used a 380-nt mog-4 probe from position −195 in the 5′-flanking region and to position +185 in the coding region and a 257-nt mog-5 probe (−92 to +171). For both genes, full-length cDNAs were obtained by joining cDNAs from the 3′ end, obtained from an oligo(dT) primed library (λRB1), with cDNAs from the 5′ end, obtained from a random-primed library (λRB2). The joining sites are AatII in mog-4 and EcoRI in mog-5. The full-length cDNA clones were cloned into the pBluescript vector (+KS, Stratagene) and sequenced by using standard procedures.
RNA Extraction and Northern Analysis.
Poly(A)+ RNA was extracted from synchronized animals as described (8). Briefly, frozen worms were homogenized in buffer containing 200 μg/ml proteinase K (Boehringer Mannheim), poly(A)+ RNA adsorbed onto oligo(dT)cellulose (Pharmacia), and eluted under low salt conditions. Typically, 2–3 μg of denatured poly(A)-enriched RNA was loaded per lane and separated on a 1.2% denaturing glyoxal gel (10). RNA was blotted on a Hybond-N filter (Amersham) and probed as described (8).
RNA Interference.
For each mog gene, a fragment of cDNA was cloned into pBluescript and used as a template for transcription with T3 or T7 RNA polymerase (Stratagene) [nt 247–799 for mog-1, nt 217–902 for mog-4, and nt 1–1451 for mog-5, with numbering starting at the initiator codon]. Similar regions were selected for other genes tested: positions 10,903–11,262 on F56D2; 23,941–24,323 on T05E8; and 38,040–38,468 on C06E1. Constructs were linearized and transcribed into cRNA according to the manufacturer's protocol (Stratagene mCAP RNA Capping kit). Antisense cRNA (1.5 μg/μl) was injected into wild-type adult hermaphrodites and progeny scored for phenotypes. Embryos not hatched >48 hr after laying were scored as dead. The time window of progeny scored starts 10 hr after injection and ends when no more embryos are produced (≈80 hr). To obtain double-stranded RNAs, strands were transcribed with T3 and T7 RNA polymerase (Stratagene), annealed, mixed equally in injection buffer (6.6 mM potassium phosphate, pH 7.3/1 mM potassium citrate, pH 7.5/0.66% polyethylene glycol 6000), and incubated for 10 min at 68°C and for 30 min at 37°C. RNA synthesis, annealing, and integrity were checked on a Tris-borate/EDTA agarose gel.
Phenotype Analysis.
Germ cells were counted in XX adults of genotype mog-4, unc-4 mog-4, mog-5, or mog-5 unc-4 mutants. Animals of 24–30 hr past L4 and raised at 20°C were stained by 4′,6-diamidino-2-phenylindole (0.5 μg/ml in ethanol) and mature sperm nuclei counted in single gonadal arms. Numbers were similar in marked and unmarked mutants.
Results and Discussion
Cloning mog-4 and mog-5.
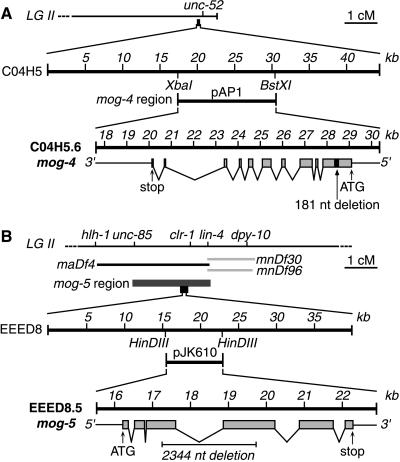
The C. elegans genome sequence (11) contains six members of the DEAH-box protein family; in addition, there are many members of the broader DEAD-box (or even more broadly DEXX-box) superfamilies, but these more divergent proteins are not considered here. One DEAH-box protein is mog-1 (8). Each of two others mapped near other mog genes: C04H5.6 maps near mog-4 and EEED8.5 near mog-5 (Fig. 1 A, Top and B, Top). To determine whether mog-4 and mog-5 might encode DEAH-box proteins, we attempted rescue of mog-4 and mog-5 homozygotes, which are normally sterile.
Figure 1.
Cloning mog-4 and mog-5. (A) mog-4. (A, Top) Genetic map of mog-4 region at right end of chromosome II (6). (A, Middle) Cosmid C04H5 resides ≈100 kb to the left of unc-52. The 12.8-kb subclone of C04H5, called pAP1, is predicted to contain only one transcript, C04H5.6. (A, Bottom) Exon/intron structure of C04H5.6, as predicted by Genefinder and confirmed by cDNA analysis (see Material and Methods). The 5′ UTR and 3′ UTR, thin lines; coding regions of exons, gray boxes; introns, lines joining exons; predicted initiation (ATG) and stop codons are indicated. The pAP2 plasmid contains a 181-nt deletion (black box) from position +582 to +763 in the mog-4 cDNA; this deletion shifts the reading frame and results in a premature stop codon. (B) mog-5. (B, Top) Genetic map of mog-5 region in the center of chromosome II (ref. 6 and this work). Genetic mapping (see Materials and Methods) placed mog-5 in a region corresponding to 2.4 cM or ≈30 cosmids on the physical map. (B, Middle) The EEED8 cosmid resides within the mog-5 region. The pJK610 subclone of EEED8 is 7.4 kb and contains only EEED8.5, the transcript predicted to encode a DEAH-box protein. (B, Bottom) Exon/intron structure of EEED8.5, as predicted by Genefinder and confirmed by cDNA analysis (see Material and Methods). Diagram represented as in Fig. 1A. The pJK611 plasmid contains a 2,344-nt deletion from position 974–2,056 in the mog-5 cDNA [or 1,200–3,544 in the mog-5 genomic fragment]; this deletion removes parts of the third and fourth exons but retains the reading frame.
We found that C04H5 and a subclone of C04H5 (pAP1) predicted to encode only one transcript, C04H5.6, both rescued mog-4(q233) homozygotes to fertility (see Materials and Methods) (Fig. 1A Middle and Bottom). Rescue was not observed with pAP2, a variant of pAP1 bearing a deletion of 181 nt from the first exon (Fig. 1A Bottom); this deletion causes a frameshift, leading to a premature stop codon. Identification of mog-4 as C04H5.6 was confirmed by sequencing the corresponding genomic DNA from mog-4(q233): this mutation is associated with a G to A transition at position 3,573 and creates a missense mutation (see below).
The results for mog-5 were parallel to those obtained for mog-4: both the cosmid EEED8 and a 7.4-kb subclone, pJK610, which is predicted to encode only EEED8.5, rescued mog-5(q449) homozygotes to fertility (see Materials and Methods). By contrast, a plasmid with a 2344-nt deletion (BspEI) that removes 361 aa from MOG-5 did not rescue mog-5(q449). The identity of mog-5 as EEED8.5 was confirmed by sequencing mog-5(q449): it is associated with a G to A transition at position 3,318, which creates a missense mutation (see below).
mog-4 and mog-5 mRNAs.
Both mog-4 and mog-5 are trans-spliced to the SL1 splice leader just before the first AUG (3 or 1 nt upstream for mog-4 and mog-5, respectively). We suggest this first AUG to be the initiator codon because it generates an N terminus conserved among close homologs for both genes (see below). The mog-4 and mog-5 cDNAs contain 3,211 and 3,731 nt, respectively, not including their poly(A) tails. At its 3′ end, mog-4 has a canonical poly(A) addition signal (AAUAAA) located 8 nt upstream of the poly(A) tail; mog-5, by contrast, possesses no well-recognized polyadenylation signal, as observed in ≈7% of C. elegans transcripts (12).
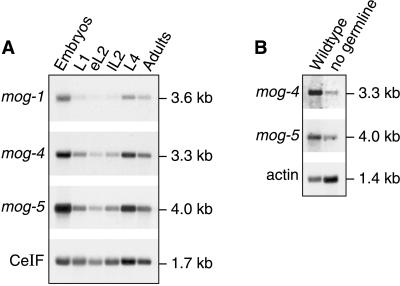
Single transcripts of expected size were observed in Northern blots when probed for mog-4 and mog-5 mRNAs (Fig. 2A). These mRNAs are abundant in embryos, low in early larval development, and increased again in later larvae and adults (Fig. 2A). We next analyzed mog-4 and mog-5 mRNAs in poly(A)+ RNA derived from either wild-type adults (which possess ≈2,000 germ cells) or glp-1 mutant adults (which have only 16–32 sperm and no other germ cells) (Fig. 2B).The mog-4 and mog-5 mRNAs were abundant in wild-type adults but reduced in glp-1 mutants. The simplest interpretation is that mog-4 and mog-5 mRNAs are expressed in the germ line as well as somatic tissues. This expression is consistent with the mog germline phenotype and with the somatic effect of mog genes in reporter assays (5). A similar expression also was observed for mog-1 (8).
Figure 2.
mog-4 and mog-5 mRNA expression. (A) mog-4 and mog-5 RNAs during development. In each lane, 3.8 μg of poly(A)-enriched RNA was loaded from embryos (E), larvae [first larval stage (L1), early L2 (eL2), late L2 (lL2), and fourth larval stage (L4)] and adults (A). Probes were the full-length cDNAs for mog-4 (pAP5) and mog-5 (pJK615), respectively. Molecular sizes were determined by comparison to Promega RNA markers. CeIF encodes the C. elegans homolog of eukaryotic initiation factor 4A, which is expressed at a constant level throughout development (28). (B) mog-4 and mog-5 RNAs are present in soma and germ line. RNA was derived from wild-type (N2) adults and glp-1(q224) adults raised at restrictive temperature, which have virtually no germ line (29). Each lane was loaded with 2.5 μg of poly(A)-enriched RNA and probed with mog-4, mog-5, and C. elegans actin-1-specific (as loading control) cDNAs. The mog-4 and mog-5 transcripts are abundant in wild-type RNA but much reduced in animals lacking a germ line. For comparison, blots were also probed with mog-1 (A and not shown).
mog-4 and mog-5 Encode DEAH-Box Proteins.
The MOG-4 and MOG-5 polypeptides contain seven conserved motifs typical of members of the DEAH-box protein family (Fig. 3, black boxes) (13, 14). Although full-length MOG-4 and MOG-5 polypeptides share only 38.4% identity, a conserved region of 617 aa (including the seven motifs) is 55.5% identical. This conserved region extends from amino acid 357–974 in MOG-4 and is demarcated by arrows in Fig. 3. We refer to this conserved region as a “domain of high similarity” and used it for phylogenetic analyses (see below). Outside this domain, MOG-5 contains multiple serine-arginine (SR) and aspartic acid-arginine (DR) dipeptides (Fig. 3, thin underline), which define an RS domain (15). Such RS regions are also found in MOG-1 and in human homologs but not in MOG-4 or in PRP2, PRP16, PRP22, or PRP43 (see below). In addition, MOG-5 contains a motif found in bacterial ribosomal protein S1 (Fig. 3, thick underline); this same motif is found in yeast PRP22 and its human homolog HRH1 (16, 17).
Figure 3.
Comparison of MOG-4 and MOG-5 amino acid sequences. Amino acids conserved between MOG-4 and MOG-5 are boxed. Amino acids in all 18 DEAH- box proteins compared in Fig. 5 are shaded black with white letters; amino acids conserved in all 16 DEAHER proteins are shaded gray (amino acids defined as conserved include: T/S, V/I/L, D/E, N/Q, R/K, F/Y, and A/G). Arrows demarcate the “domain of high similarity.” The RS domain in MOG-5 is indicated by a thin underline; the S1 domain in MOG-5 is shown by a thick underline. Altered amino acids in mog-4(q233) and mog-5(q449) mutant alleles are indicated by ○ and ●, respectively.
A DEAHER Subfamily of DEAH-Box Proteins.
Using the blastp program, 15 proteins were identified as well-matched to MOG-1, MOG-4, and MOG-5: three additional proteins from C. elegans (F56D2.6, C06E1.10, and T05E8.3), four from Saccharomyces cerevisiae (PRP2, PRP16, PRP22, and PRP43), one from Schizosaccharomyces pombe (Cdc28), five from vertebrates [KIAA0057, hPRP16, HRH1, mDEAH9 (18), and DBP1 (19)], and two from Arabidopsis thaliana (helicases 1 and 2; GenBank accession nos. X981340 and Z97341). A comparison of the “domains of high similarity” of these 18 proteins revealed features that were well conserved among a subset of 16 proteins (all but C06E1.10 and T05E8.3). Whereas all 18 possess the groups of amino acids shared by all DEAH-box proteins (Fig. 3, black boxes), the subset of 16 also share an additional 120 amino acids scattered throughout the “domain of high similarity” (Fig. 3, gray boxes). Among these are amino acids flanking the more broadly conserved amino acids: following DEAH is ER(T/S) in motif III, and preceding PRRVAA is TQ in motif Ia (already noted in ref. 14). In addition, the 16 share a common size in their domains of high similarity [the shortest is 593 aa (KIAA0057) and the longest is 640 aa (PRP2)]. The T05E8.3 protein shares 116/120 aa marked in gray (Fig. 3) and has several insertions into its domain of high similarity. Remaining homologs, including C. elegans C06E1.10, other DEAH proteins, such as the Bloom syndrome RecQ helicases (ref. 20 and for review see ref. 21), and other helicases belonging to the broader DEXH family [e.g., the Drosophila Homeless and Maleless proteins (22, 23)], are even further diverged. We propose that the shared features revealed by this comparison will prove useful and suggest the name DEAHER-box proteins to distinguish this class of proteins.
Critical Amino Acids in DEAHER-Box Proteins.
The mog-4 and mog-5 mutations each alter a conserved residue. In mog-4(q433), a conserved glycine (G) is changed to a serine (S) (Fig. 3, ○) in motif VII (QRXGRAGR), a motif involved in ATP hydrolysis in the DEAD-box protein eIF-4A (13). In yeast PRP16, an alanine substitution of glycine 691, which corresponds to the altered amino acid in mog-4(q233), is not essential (24); however, the same amino acid is critical for catalytic activity of the vaccinia virus NPH-II DEVH-box protein (25). Although a change to serine might be more detrimental than one to alanine, mog-4(233) is probably not a null allele (see below). In mog-5(q449), a conserved glutamic acid (E) is changed to lysine (K) (Fig. 3, ●). Furthermore, in a prp22 temperature-sensitive allele, a conserved glycine, which corresponds to amino acid 659 in MOG-4, is changed to glutamic acid (16). Finally, mog-1(q473) changes a conserved arginine [R702 in MOG-4] to histidine (H) (8). Therefore, all known missense mutations in the mog genes and close homologs alter conserved amino acids.
Null and Non-Null Mog Phenotypes.
Among the four mog-1 alleles examined previously, three were nonsense mutants predicted to be null, and one was a missense mutant (8). Intriguingly, the mog-1 nonsense mutants were phenotypically distinguishable from the mog-1(q473) missense mutant. Both types of mutant failed in their switch to oogenesis, but the null mutants had an additional defect in germline proliferation (8). We have found that the germ lines of mog-4(q233) and mog-5(q449) missense mutants are comparable to that of the mog-1 missense mutant with respect to germline proliferation: 1,073 ± 341 (n = 18) (8), 638 ± 173 (n = 12), or 775 ± 216 (n = 10) sperm per arm in mog-1(q473), mog-4(q233), and mog-5(q449), respectively. By contrast, mog-1 null mutants possessed ≈300 sperm per arm. The total germ cells (immature plus gametes) was also larger in the missense mutants than in the mog-1 null mutants. We therefore suggest that the mog-4(q233) and mog-5(q449) mutants may retain partial activity and that null alleles of mog-4 or mog-5 may cause a different phenotype, for instance embryonic or larval lethality.
Comparison of C. elegans DEAH-Box Proteins by RNA Interference.
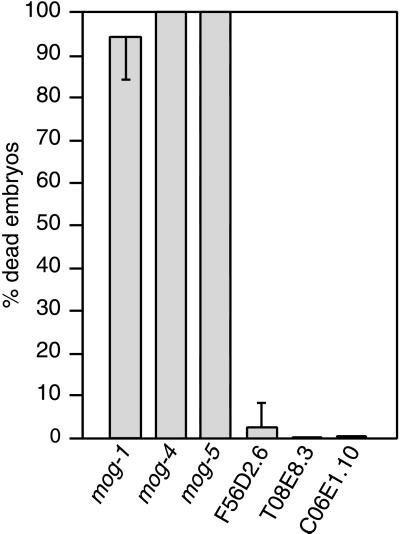
To compare functions of the C. elegans DEAH-box and DEAHER proteins, we used RNA interference (RNAi) to reduce gene function. For mog-1, mog-4, and mog-5, RNAi resulted in embryonic lethality (Fig. 4), which was expected: the mog genes are required maternally for embryogenesis (6, 7). Occasionally, embryos laid early after injection developed into sterile adults making only sperm. The mog(RNAi) embryos arrested at specific stages: mog-1(RNAi) embryos arrested with ≈300 cells at the early comma stage, whereas mog-4(RNAi) and mog-5(RNAi) embryos arrested earlier (166 ± 9 cells for mog-4 and 130 ± 10 cells for mog-5) (not shown).
Figure 4.
RNA interference against C. elegans genes encoding DEAH-box proteins. Embryonic lethality observed after RNA interference directed against six C. elegans genes encoding DEAH-box proteins. Progeny were scored for arrested embryos 24 hr after eggs were laid. mog-1, n = 756; mog-4, n = 600; mog-5, n = 276; F56D2.6, n = 629; C06E1.10, n = 929; and T05E8.3, n = 752.
RNAi directed against the three remaining C. elegans DEAH- box proteins resulted in little or no embryonic lethality (Fig. 4). This same result was observed with either single-stranded (ss) antisense RNA (Fig. 4) or double stranded (ds) RNA (not shown). Furthermore, neither ssRNAi or dsRNAi progeny exhibited a Mog germ line. F56D2.6(dsRNAi) and C06E1.10(dsRNAi) progeny grew more slowly than normal and T05E8.3 had no obvious defect (not shown). We also compared the map positions of these DEAH-box genes to the other mog genes. The mog-2 and mog-6 genes map to chromosome II, whereas mog-3 maps to the left of dpy-17 on chromosome III (ref. 6 and A.P. unpublished data). By contrast, F56D2.6 maps to the right of dpy-17 on chromosome III, T05E8.3 maps to chromosome I, and CO6E1.10 maps to the center of chromosome III. Therefore, these three DEAH-box proteins do not map to any known mog gene and do not exhibit either the embryonic lethality or defective germline phenotypes typical of mog mutants. We suggest that MOG-1, MOG-4, and MOG-5 define a functional subset of DEAH-box proteins within the C. elegans genome.
Phylogenetic Analysis of DEAH-Box Proteins.
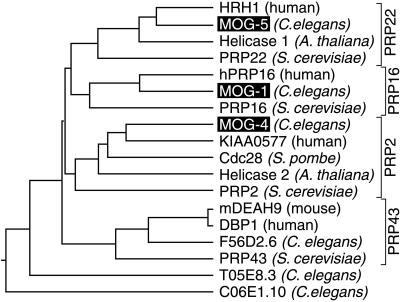
To investigate the evolutionary relationships among the 18 most similar DEAH-box proteins of different species, we compared their “domains of high similarity” (Fig. 3, region between arrows) using the clustal method (DNAstar) and GCG pileup programs (Fig. 5; not shown). This analysis identified four clusters for the DEAHER subset of DEAH-box proteins, each containing a well-characterized PRP protein from S. cerevisiae (Fig. 5). The only proteins not falling into one of these clusters were T05E8.3 and C06E1.10 (Fig. 5), which also did not conform to the DEAHER family definition. Two additional C. elegans proteins, T07D4.3 and F52B5.3, are much more diverged and beyond the scope of this work.
Figure 5.
Phylogenetic tree of DEAH-box proteins. Neighbor joining phylogram of the conserved region of 18 DEAH-box proteins. The region used to generate this phylogram lies between the two arrows in Fig. 3. The phylogram was obtained by the clustal method in the DNAstar DNA analysis package. Similar results were obtained with an uncorrected distance matrix using the pileup algorithm (GCG, Madison, WI).
The clustering of DEAHER proteins into four groups, which was based on their domains of high similarity (Fig. 3, region between arrows), is supported by similarities in other parts of the proteins as well. Unique to the PRP16 group is the DR[E/D]WY[D/M][N/M][D/E] motif (at position 230 and 171 in MOG-1 and PRP16, respectively) (8). Distinctive to the PRP22 group is a conserved S1 domain in the N terminus (Fig. 4, thick underline). A shared feature of the PRP43 group is their short overall length: PRP43, mDEAH9, DBP1, and F56D2.6 have 767, 758, 813, and 739 amino acids respectively versus averages of 1171, 1133, and 955 for the PRP22, PRP16, and PRP2 groups. Although no specific motif or unique feature was discerned for the PRP2 group, MOG-4 was closely related to KIAA0057 (both lack an RS domain, have 68.8% identity in the region of high similarity and 53.9% identity throughout the entire peptide).
We conclude that MOG-1 is closely related to PRP16, MOG-4 to PRP2, and MOG-5 to PRP22. Indeed, a search of the entire C. elegans genome reveals no better homologs. The idea that MOG-1, MOG-4, and MOG-5 are the C. elegans homologs of yeast proteins PRP16, PRP2, and PRP22, respectively, is supported by comparison with the human homologs hPRP16 and HRH1, which are even more similar to the C. elegans proteins than to the proposed yeast homologs (17, 26). We tried to rescue prp16 and prp22 mutants with the respective C. elegans mog-1 and mog-5 cDNAs, but were unsuccessful (A.P. unpublished data). This failure to complement the yeast mutants was, however, predictable because rescue of prp16 with hPRP16 was only possible with a human-yeast chimeric gene (26). Similarly, HRH1 was barely able to complement a temperature-sensitive prp22 mutant strain at 32°C (17).
In yeast, PRP16, PRP2, and PRP22 are integral components of the spliceosomal machinery (27). However, in C. elegans, the biochemical functions of MOG-1, MOG-4, and MOG-5 remain unknown. Although a role for the mog genes in splicing remains possible, no general splicing defect was observed in mog-1 null mutants (8). Furthermore, the mog genes are required for PME-repression of reporter transgenes (5). One explanation of these apparently disparate results is that the mog genes, although evolutionarily related to the yeast PRP genes, have acquired a different function. One speculative idea is that the MOG proteins may promote a conformational change or the disassembly of a ribonucleoprotein complex required for PME-mediated repression. Alternatively, the PRP16, PRP2, and PRP22 proteins in yeast may function more broadly than previously thought and modulate a variety of ribonucleoprotein complexes, including those integral to splicing.
Acknowledgments
We are grateful to Jim Dahlberg, Phil Anderson, Betsy Goodwin, and Eric Haag for comments on the manuscript. We thank the C. elegans Sequencing Consortium for providing C. elegans genome sequence before publication and Alan Coulson for cosmids. We are grateful to R. Barstead for cDNA libraries and to the Caenorhabditis Genetics Center and Theresa Stiernagle for C. elegans strains. We thank Laura Vanderploeg for figure preparation. A.P. was supported by the European Molecular Biology Organization and the Howard Hughes Medical Institute. J.K. is an investigator with the Howard Hughes Medical Institute. This work was in part supported by Swiss National Science Foundation Grants 823A-042914, 3100–055384.98/1, and 3130–054989.98/1 (to A.P.).
Abbreviations
- UTR
untranslated region
- PME
point mutation element
- fem
feminization
- mog
masculinization of the germ line
- PRP
pre-mRNA processing
- RNAi
RNA-mediated interference
References
- 1.Schedl T. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 241–269. [PubMed] [Google Scholar]
- 2.Hodgkin J. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton M K, Schedl T B, Kimble J. Genetics. 1987;115:107–119. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahringer J, Kimble J. Nature (London) 1991;349:346–348. doi: 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- 5.Gallegos M, Ahringer J, Crittenden S, Kimble J. EMBO J. 1998;17:6337–6347. doi: 10.1093/emboj/17.21.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham P L, Schedl T, Kimble J. Dev Genet (Amsterdam) 1993;14:471–484. doi: 10.1002/dvg.1020140608. [DOI] [PubMed] [Google Scholar]
- 7.Graham P L, Kimble J. Genetics. 1993;133:919–931. doi: 10.1093/genetics/133.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puoti A, Kimble J. Mol Cell Biol. 1999;19:2189–2197. doi: 10.1128/mcb.19.3.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly W G, Xu S, Montgomery M K, Fire A. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 11.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 12.Blumenthal T, Steward K. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 117–145. [Google Scholar]
- 13.Pause A, Sonenberg N. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arenas J E, Abelson J N. Proc Natl Acad Sci USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu X-d. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 16.Company M, Arenas J, Abelson J. Nature (London) 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 17.Ono Y, Ohno M, Shimura Y. Mol Cell Biol. 1994;14:7611–7620. doi: 10.1128/mcb.14.11.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gee S, Krauss S W, Miller E, Aoyagi K, Arenas J, Conboy J G. Proc Natl Acad Sci USA. 1997;94:11803–11807. doi: 10.1073/pnas.94.22.11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamura O, Sugawara W, Furuichi Y. Biochem Biophys Res Commun. 1997;240:335–340. doi: 10.1006/bbrc.1997.7585. [DOI] [PubMed] [Google Scholar]
- 20.Ellis N A, Groden J, Ye T Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 21.Kusano K, Berres M E, Engels W R. Genetics. 1999;151:1027–1039. doi: 10.1093/genetics/151.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroda M I, Kernan M J, Kreber R, Ganetzky B, Baker B S. Cell. 1991;66:935–947. doi: 10.1016/0092-8674(91)90439-6. [DOI] [PubMed] [Google Scholar]
- 23.Gillespie D E, Berg C A. Genes Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- 24.Hotz H-R, Schwer B. Genetics. 1998;149:807–815. doi: 10.1093/genetics/149.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross C H, Shuman S. J Virol. 1996;70:1706–1713. doi: 10.1128/jvi.70.3.1706-1713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Z, Reed R. EMBO J. 1998;17:2095–2106. doi: 10.1093/emboj/17.7.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burge C B, Tuschl T, Sharp P A. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. [Google Scholar]
- 28.Roussell D L, Bennett K L. Nucleic Acids Res. 1992;20:3783. doi: 10.1093/nar/20.14.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Austin J, Kimble J. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]